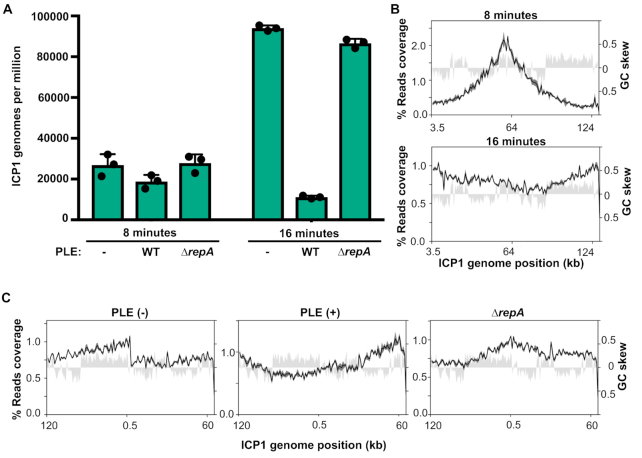
Figure 8.
Non-replicating PLE still alters ICP1’s replication profile. (A) Genomes per million of ICP1 in PLE(−), PLE(+), and PLE ΔrepA cultures at 8 and 16 min post-infection. Values shown are the means of three biological replicates. (B) Percent reads coverage profile of ICP1’s genome in ΔrepA PLE infection at 8 min (top) and 16 min (bottom) post-infection. (C) Percent reads coverage profile of ICP1’s genome in PLE (−), PLE (+), and ΔrepA PLE hosts at 16 min post infection. The ICP1 genome has been rotated so that it is centered around the putative rolling circle replication origin. For each reads profile plot in (B) and (C), the -average percent reads coverage across the genome for three biological replicates is shown as a black line, while standard deviation appears as dark gray shading around the line. The GC skew (right axis) is plotted as light gray shading.