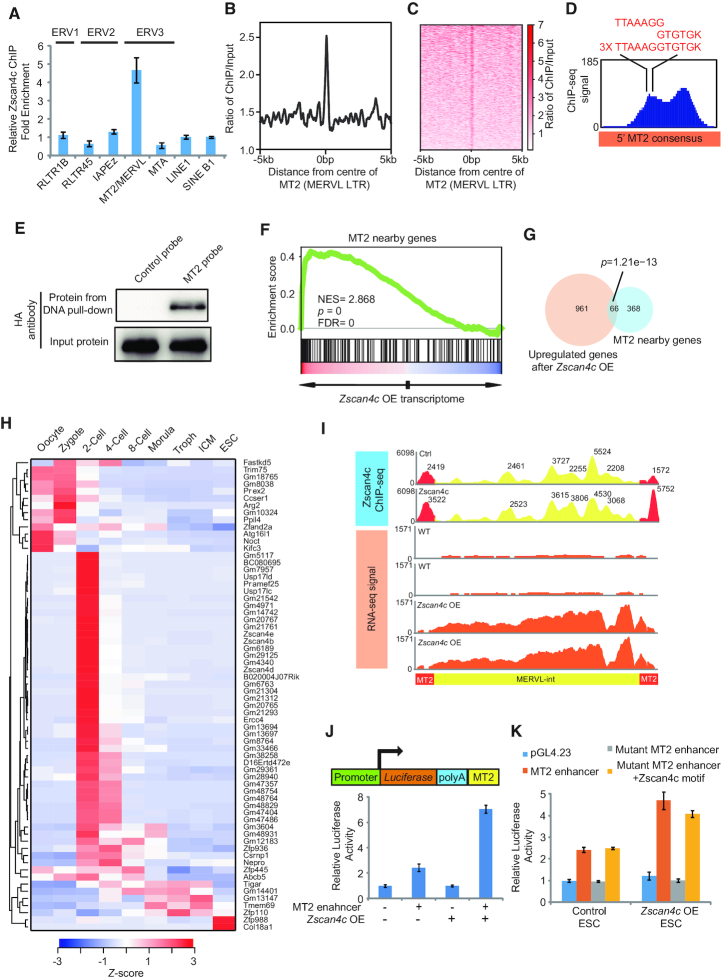
Figure 5.
Zscan4c interacts with MT2 and regulates nearby genes. (A) ChIP-qPCR analysis of Zscan4c binding on different retroelements. ChIP-qPCR data was normalized to input and that of LINE1. Biological triplicate data (n = 3 extracts) are presented as mean ± s.e.m. (B, C) Zscan4c binding profile plot (B) and enrichment heatmap (C) around the centre of MT2 sequence. The ChIP-seq signal was calculated as the ratio of normalized reads relative to input. (D) Zscan4c binding signal on MT2 and position of Zscan4c motif on MT2. Red sequences highlight the sequence of Zscan4c binding motif identified on MT2. (E) DNA pull-down analysis of the DNA binding activity of Zscan4c. DNA pull-down was done with 293T cells overexpressing HA-tagged Zscan4c. Probe with scrambled sequence was used as control. (F) MT2 nearby genes were enriched in upregulated genes after Zscan4c overexpression. Genes within 10 kb from MT2 locus were considered to be near MT2. Red, upregulated genes; blue, downregulated genes. (G) Overlap between upregulated genes after Zscan4c overexpression and MT2 10 kb nearby genes. P value was calculated by Fisher's exact test. (H) Expression heatmap of overlapped genes between upregulated genes after Zscan4c overexpression and genes within 10 kb from MT2. Troph, trophectoderm; ICM, inner cell mass. Average expression level of each developmental stage was determined and presented as Z-score across different stages. (I) Density plot of Zscan4c ChIP-seq signal and RNA-seq expression of MT2/MERVL consensus sequence from GeneBank (ID: Y12713.1). Density plot was plotted by IGV with ChIP-seq and RNA-seq bam files. Yellow region, ChIP-seq signal on MERVL-int; red region, ChIP-seq signal on MT2. ChIP input was included as control. Value of individual peak was highlighted. (J) Luciferase assay analysis of enhancer activity of MERVL LTR(MT2) after Zscan4c overexpression. MT2+, MT2 at the downstream of Luciferase gene; MT2–, reporter without MT2. Zscan4c OE +, Zscan4c overexpression; Zscan4c OE–, control vector overexpression. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m. (K) Luciferase assay analysis enhancer activity of wild type MT2, MT2 with mutant Zscan4c binding motif, and mutant MT2 with inserted Zscan4c binding motif after Zscan4c overexpression. Biological-triplicate data (n = 3 extracts) are presented as mean ± s.e.m.