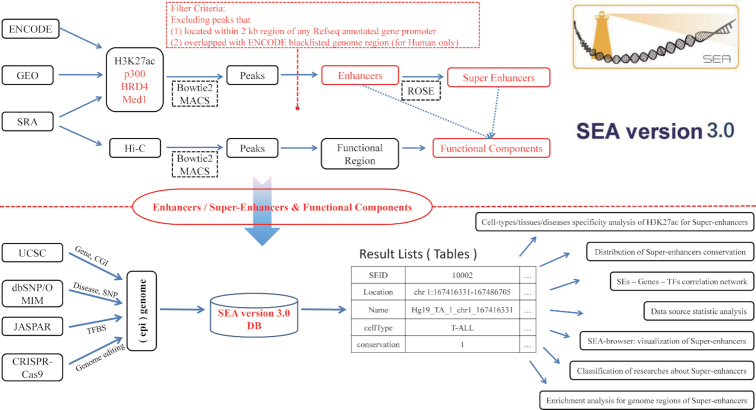
Figure 1.
Database content and construction. SEA v. 3.0 takes advantage of available public H3K27ac, BRD4, Med1 and p300 ChIP-Seq datasets to identify super-enhancers in different cell types/tissues/diseases of 11 species. It excludes peaks located within ±2 kb of any RefSeq annotated gene promoter or peaks overlapping with ENCODE blacklisted genomic regions. Multiple track types are used for genomic visualization including functional components generated by Hi-C datasets. Shannon Entropy is used to calculate and evaluate the cell type specificity of super-enhancers, and all data are accessible through the download page.