Figure 2.
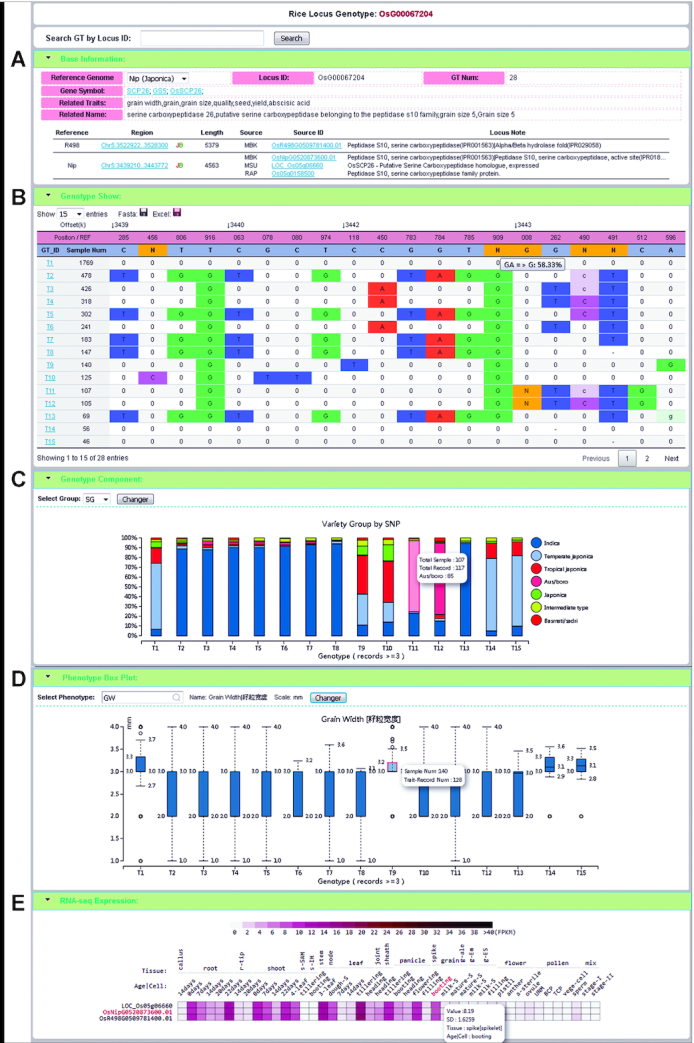
An example of a locus page showing multiple types of information associated with the locus. Rice gene GS5 controlling grain size is on locus OsG00067204. (A) Two reference genomes contain different alleles at the locus. (B) Genotype table showing all genotypes in the rice population, with reference genome positions in the first row of the table and reference bases in the second row. Genotype IDs are enumerated from T1 as the most frequent genotype (allele or haplotype for homozygous genome). ‘0’ indicates that the genotype at this position is the same as in the reference genome. Capitalized bases indicate homozygous variants, lowercase bases indicate heterozygous variants and ‘-’ indicates missing information. In the REF row, the letter ‘N’ represents a short insertion in the reference genome compared with some other samples. In the GT row, the ‘N’ represents insertion of bases in the samples compared with the reference genome. (C) Distribution of each genotype (allele) in a rice population. For example, the majority of genotype (allele) T1 is found in temperate japonica lines (1251 out of 1769). (D) Boxplot showing the relationship between genotypes and phenotypes. The phenotype can be selected by users from all collected traits in the database. In this case, the locus OsG0067204 is known to be associated with grain width (GW) traits. (E) Expression profile at the locus in different reference genomes in different tissues.
