Figure 5.
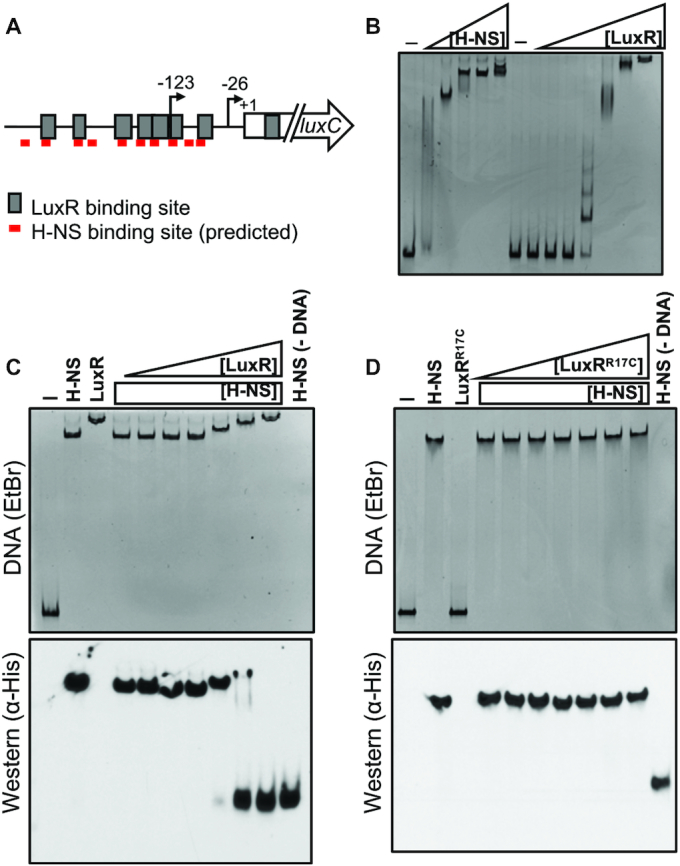
H-NS is displaced from PluxC DNA by LuxR in vitro. (A) Diagram of luxCDABE promoter. Gray boxes indicate LuxR binding sites, and red lines indicate FIMO-predicted H-NS binding sites. Arrows indicate transcription start sites relative to the start codon (+1). (B) EMSA reactions containing 8.8 nM PluxC DNA and either H-NS (100, 150, 200, 250, 300 nM) or LuxR (31.25, 62.5, 125, 250, 500, 1000, 2000 nM) purified protein. Reactions denoted ‘–’ contained no protein. (C) Competitive EMSA reactions consisting of 8.8 nM PluxC DNA and 200 nM purified H-NS and either 31.25, 62.5, 125, 250, 500, 1000 or 2000 nM purified LuxR. Lanes labelled ‘H-NS’, ‘LuxR’, ‘–’, and H-NS (–DNA) contained 200 nM H-NS, 2000 nM LuxR, no protein, and 200 nM H-NS without DNA, respectively. The reactions were run on polyacrylamide gels and stained with ethidium bromide (top) and then transferred to a nitrocellulose membrane and probed for H-NS using α-His-HRP antibodies (bottom). (D) The same experimental procedures were used as described in panel C but instead using the DNA-binding deficient LuxRR17C purified protein.
