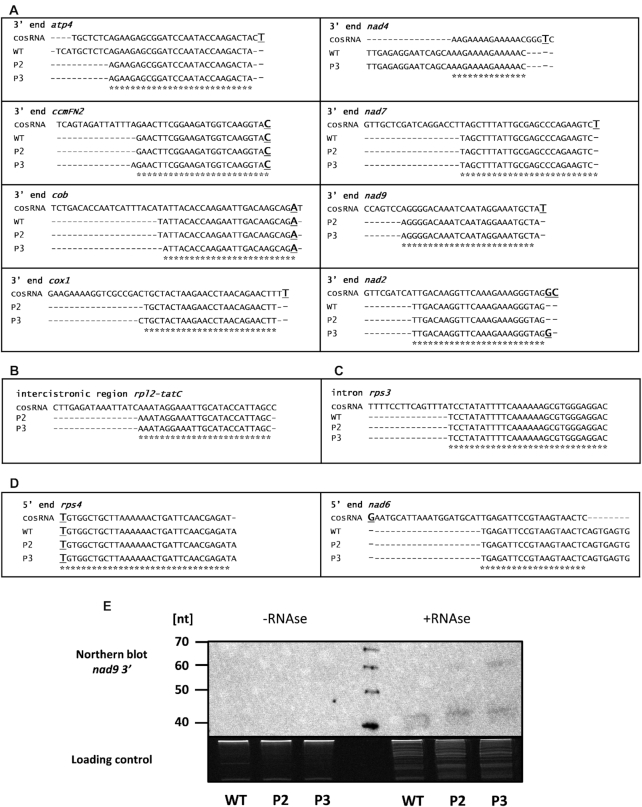
Figure 4.
mtRibo-seq-sRNAs identified in P2 and P3 phenotypes of rps10 and wild-type. Comparison of mtRibo-seq-sRNAs sequences identified in mtRibo-seq reads in rps10 and wild-type plants with previously published cosRNAs (A) from 3′ ends of mRNAs, (B) intercistronic regions, (C) introns and (D) 5′ ends of mRNAs. The cosRNA sequences are from (18). Asterisks mark sequences common to mtRibo-seq-sRNA and cosRNA. Major mRNA ends are in bold and underlined. (E) Detection of nad9 mtRibo-seq-sRNA in sRNA fractions of mitochondrial polysomal RNA isolated from rps10 and wild-type plants. Equal amounts (500 ng) of two sRNA fractions were analyzed: sRNAs isolated from polysomal fraction, and sRNAs isolated from polysomal fraction treated with RNase before isolation of sRNAs. The fractions were run on a denaturing 15% polyacrylamide gel, transferred to a nylon membrane and detected using 5′- biotinylated oligonucleotide probe. The image of the agarose gel stained with ethidium bromide is shown as a loading control in the bottom panel.