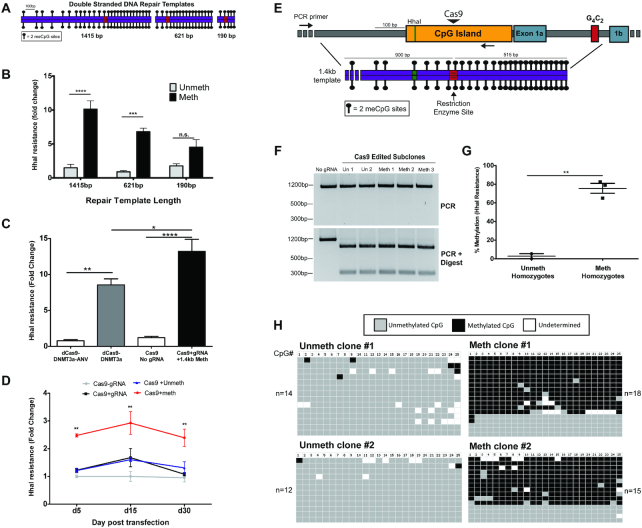
Figure 2.
Editing DNA methylation with double-stranded repair templates. (A) Diagram of double stranded DNA repair templates (either fully methylated or fully unmethylated at CpG sites) homologous to the C9orf72 promoter. Red box indicates a 2 bp substitution that generates a restriction enzyme site and mutates the PAM sequence. Figure drawn to scale. (B) MSRE-qPCR assay to measure DNA methylation using a long amplicon that anneals outside the repair templates. HEK293T cells were transfected with CRISPR/Cas9+gRNA and the indicated templates. Cells were puromycin selected for 2 days and collected at day 4 post transfection. n = 3 experiments; two-way ANOVA shows significant interaction between template length and template methylation status (P = 0.0071). Bonfferoni post-hoc test between unmeth and meth template groups ****P < 0.0001, *** P < 0.001. (C) Comparison of methylation efficiency between HARDEN method and dCas9-DNMT3a fusion protein construct. HEK293T cells were transfected with indicated plasmids and methylation was measured 2 days post transfection using long amplicon MSRE-qPCR. n = 3 experiments. One-way ANOVA (P < 0.0001) followed by Bonferroni post-hoc comparison of indicated groups. ****P < 0.0001, **P < 0.01, *P < 0.05. (D) Timecourse of methylation measured via long amplicon MSRE-qPCR using 1.4kb dsDNA repair templates. n = 3 experiments. Two-way ANOVA (Group P < 0.0001; day post transfection P = 0.0343) followed by Bonferroni post-hoc comparison between unmethylated template and methylated template groups. **P < 0.01. (E) Diagram of C9orf72 promoter region indicating the position of the repair template used to generate stably methylated HEK293T clones. Red box indicates a 2 bp substitution that generates a restriction enzyme site and mutates the PAM sequence. Figure drawn to scale. (F) PCR and digest of HEK293T clonal lines that were repaired using the 1.4kb dsDNA repair templates. Cells were collected ∼5 weeks post transfection. (G) MSRE-qPCR of clonal cell lines that were homozygous for editing with either the unmethylated (n = 2) or methylated 1.4kb repair template (n = 3). Cells were collected ∼5 weeks post transfection. Two-tailed t-test; P = 0.0020. (H) Bisulfite amplicon sequencing of homozygous clonal cell lines edited with the unmethylated or methylated 1.4kb repair templates. Cells were collected ∼5 weeks post transfection. Each row is an individual DNA molecule and columns are individual CpG dinucleotides. Grey boxes indicate unmethylated CpGs, black boxes indicate methylated CpGs and white boxes are undetermined due to poor conversion or poor sequencing quality.