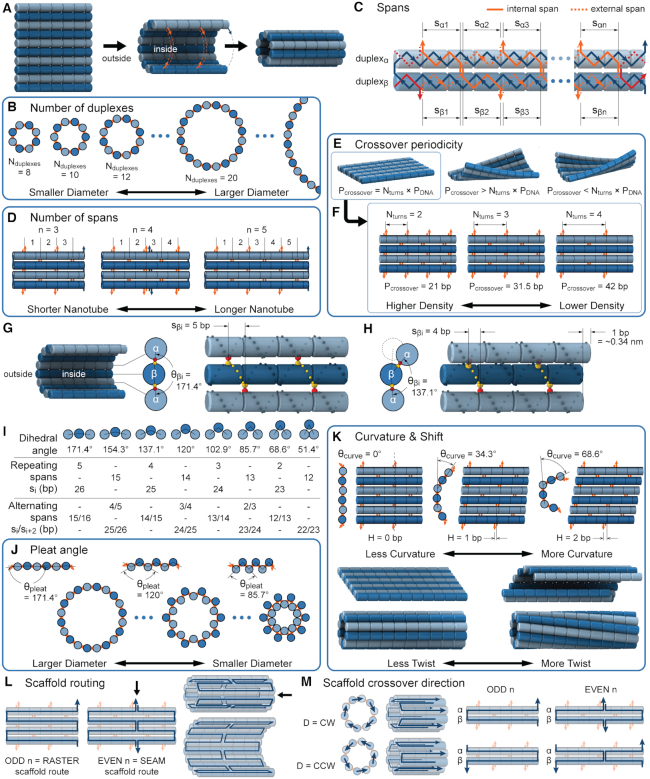
Figure 2.
Description of DNA origami nanotube design parameters. (A) A reference DNA origami sheet with an arbitrarily defined inside and outside surface, and the resulting nanotube when top and bottom helices are linked by crossovers. (B) Along-axis view of nanotubes with different even numbers of duplexes (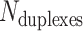 ). (C) A neighbouring pair of DNA duplexes (
). (C) A neighbouring pair of DNA duplexes (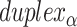 and
and 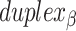 ), which are repeated around the circumference of the nanotube. Their associated list of staple spans (
), which are repeated around the circumference of the nanotube. Their associated list of staple spans (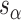 and
and 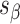 ), which quantify the number of base pairs between successive staple crossovers are labelled and shown as solid orange lines for internal spans or dotted orange lines for external spans. End-spans are shown as red lines and occur between the scaffold crossover at the end of the nanotube and the proximal staple crossover. (D) DNA origami sheets with different number of spans (
), which quantify the number of base pairs between successive staple crossovers are labelled and shown as solid orange lines for internal spans or dotted orange lines for external spans. End-spans are shown as red lines and occur between the scaffold crossover at the end of the nanotube and the proximal staple crossover. (D) DNA origami sheets with different number of spans (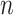 ), which affects the length of the nanotube. (E) DNA origami sheets each with a different crossover periodicity (
), which affects the length of the nanotube. (E) DNA origami sheets each with a different crossover periodicity (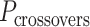 ). On the left,
). On the left, 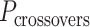 is equal to the periodicity of DNA (
is equal to the periodicity of DNA (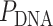 ) multiplied by an integer number of complete turns (
) multiplied by an integer number of complete turns (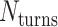 ) resulting in a flat sheet. Whereas the middle and right depict DNA origami sheets where
) resulting in a flat sheet. Whereas the middle and right depict DNA origami sheets where 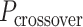 is out of phase with
is out of phase with 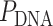 in opposite directions resulting in a global twist also in opposite directions. (F) DNA origami sheets with different values of
in opposite directions resulting in a global twist also in opposite directions. (F) DNA origami sheets with different values of 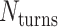 where the density of staple crossovers (orange) is inversely proportional to
where the density of staple crossovers (orange) is inversely proportional to 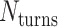 . (G) Three adjacent DNA duplexes with an internal span length of
. (G) Three adjacent DNA duplexes with an internal span length of 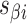 = 5 bp resulting in a dihedral angle of
= 5 bp resulting in a dihedral angle of 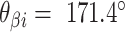 relative to the inside of the DNA sheet. Red and yellow spheres show the midpoint between successive C3′ carbon atoms (NEMid) (59) on
relative to the inside of the DNA sheet. Red and yellow spheres show the midpoint between successive C3′ carbon atoms (NEMid) (59) on 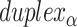 and
and 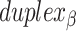 respectively (Supplementary Figure S1). (H) Three adjacent DNA duplexes with an internal span length of
respectively (Supplementary Figure S1). (H) Three adjacent DNA duplexes with an internal span length of 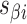 = 4 bp resulting in a dihedral angle of
= 4 bp resulting in a dihedral angle of 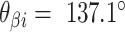 and a shift of 1 bp. (I) Table of the eight possible dihedral angles used in this study (including the seven angles between 60° and 180°) and the possible repeated or alternating span lengths required to achieve them. (J) Illustration of how the pleat angle (
and a shift of 1 bp. (I) Table of the eight possible dihedral angles used in this study (including the seven angles between 60° and 180°) and the possible repeated or alternating span lengths required to achieve them. (J) Illustration of how the pleat angle (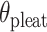 ) of a sheet or nanotube is defined as the effective dihedral angle of every
) of a sheet or nanotube is defined as the effective dihedral angle of every 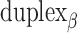 , which is proportional to sheet width and nanotube diameter. (K) DNA origami sheets with different curvature angles (
, which is proportional to sheet width and nanotube diameter. (K) DNA origami sheets with different curvature angles (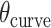 ) and associated shift values (
) and associated shift values (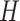 ). Bottom shows how sheets with intrinsic curvature will also be chiral, and hence are predicted to form chiral tubes. (L) DNA sheet showing scaffold strand following a raster (left) or seamed (middle) routing, where staple crossovers form a seam in the middle of the tube (indicated by arrow in middle panel). In a seamed configuration, two of the duplexes are not connected by scaffold crossovers (arrow in right panel), which may allow the formation of ‘open’ structures. (M) Depiction of nanotubes with different scaffold strand configurations. Top left and bottom left shows nanotubes with scaffold strand crossovers routed in a clockwise (CW) or counter-clockwise (CCW) direction on the left of the nanotube. Middle and right shows nanotubes with an odd and even number of spans (n), which results in a raster and seamed scaffold routing respectively.
). Bottom shows how sheets with intrinsic curvature will also be chiral, and hence are predicted to form chiral tubes. (L) DNA sheet showing scaffold strand following a raster (left) or seamed (middle) routing, where staple crossovers form a seam in the middle of the tube (indicated by arrow in middle panel). In a seamed configuration, two of the duplexes are not connected by scaffold crossovers (arrow in right panel), which may allow the formation of ‘open’ structures. (M) Depiction of nanotubes with different scaffold strand configurations. Top left and bottom left shows nanotubes with scaffold strand crossovers routed in a clockwise (CW) or counter-clockwise (CCW) direction on the left of the nanotube. Middle and right shows nanotubes with an odd and even number of spans (n), which results in a raster and seamed scaffold routing respectively.