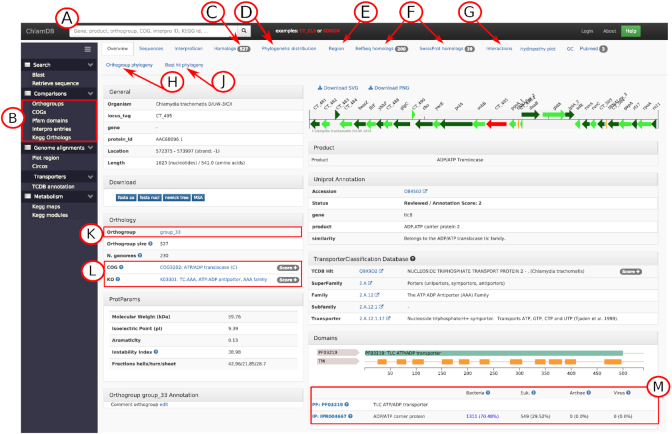
Figure 1.
Protein annotation page of CT_495, an ADP/ATP transporter. (A) Main search bar. (B) Menu to access comparative analyses tools for the comparison of genome content based on the clustering of proteins into orthologous groups and for comparison of COG, KEGG, Pfam and InterPro annotations. (C) ‘Homologs’ tab with the list of the 527 orthologs of CT_495 in other PVC genomes. (D) Tab with the reference species phylogeny and the pattern of presence/absence of orthologs in each genome of the database as well as the locus tag of the closest ortholog in each genome. (E) ‘Region’ tab showing the conservation of proteins encoded in the direct neighborhood of the target protein. (F) Best hits in RefSeq and SwissProt databases. (G) Predicted protein interactors based on phylogenetic profiling and conservation of gene neighborhood. (H) Phylogenetic trees of the orthogroup and associated Pfam and transmembrane (TM) domain organization of each protein. (J) Phylogenetic tree including the best RefSeq hits of each protein of the orthologous group. (K) Name of the orthologous group with link to an overview of the annotation of the considered orthogroup (here 527 orthologs for group_33). (L) COG and KEGG annotations with link to the detailed list of proteins annotated with the same COG/KO in other genomes of the database; (M) Pfam and InterPro annotations with basic taxonomic information from the InterPro website: the numbers and percentages of proteins harboring this domain that are classified as Bacteria, Eukaryote, Archaea and Virus (data retrieved from InterPro version 60). Clicking on the Pfam accession numbers links to more detailed taxonomic information and a detailed list of proteins harboring the same domain in 6677 representative RefSeq genomes.