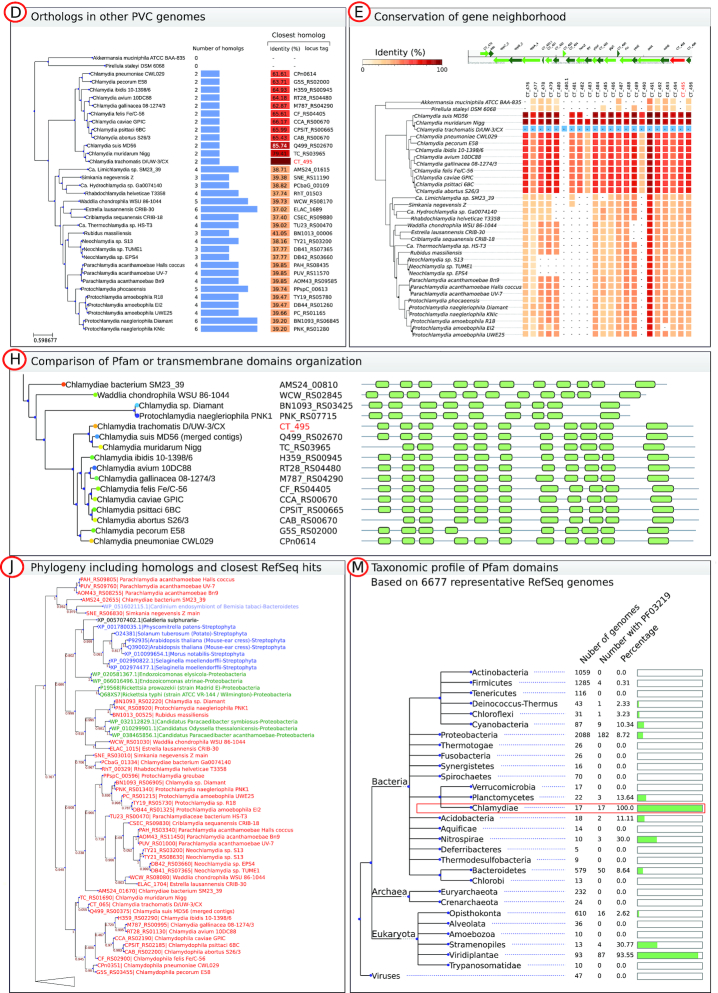
Figure 2.
Selected examples of comparative data that can be retrieved from protein annotation pages. Panels are named according to the links shown in Figure 1. (D) Profile of presence/absence of orthologs in other PVC genomes with the identity of the closest ortholog in each genome. (E) Visualization of the conservation of proteins encoded in the neighborhood of CT_495. (H) Comparative view of transmembrane domains organization of CT_495 (TlcB) orthologs. (J) Phylogeny of CT_495 orthologous group including the closest identified RefSeq homologs. Red labels are proteins from the ChlamDB database whereas blue and green labels indicate non-PVC proteins. In this example, a sequence of Cardinium, a Bacteroidetes endosymbiont of the whitefly Bemisia tabaci, is clustering with Chlamydiae spp. and Proteobacteria symbionts such as Paracaedibacter and Rickettsia spp. are clustering with other Chlamydia spp., suggesting multiple events of horizontal gene transfer. Panels present only a subset of the 277 genomes currently present in the database to fit on a single page. Complete figures can be retrieved from the ChlamDB website. (M) Overview of the taxonomic profile of the Pfam domain PF03219 in 6677 representative RefSeq genomes. We can observe that this domain can be found in 100% of Chlamydiae genomes and in most Viridiplantae, but also in some genomes of other bacterial phyla. The detailed list of hits can also be browsed and downloaded.