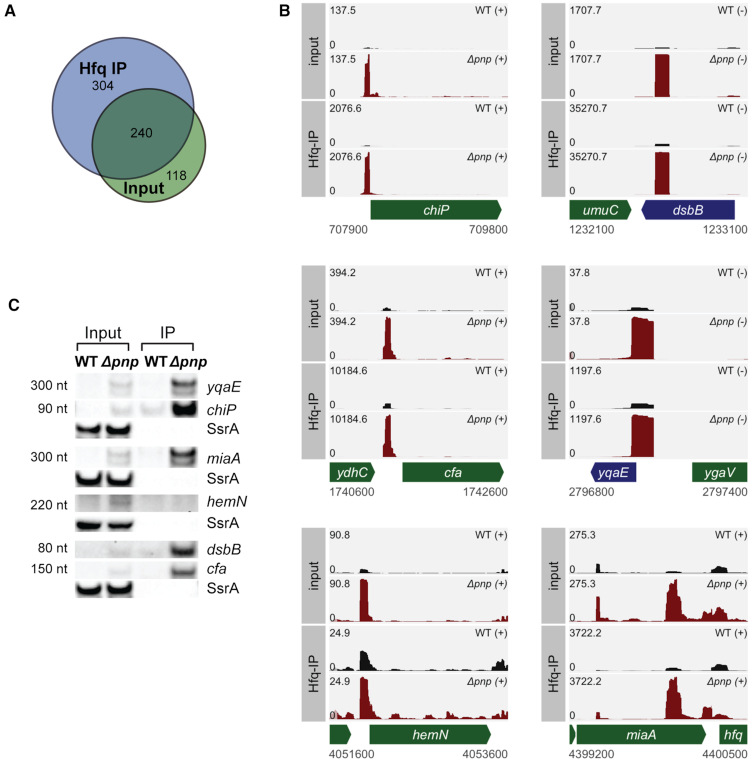
Figure 2.
mRNA-derived fragments that accumulate in the absence of PNPase interact with Hfq. (A) A Venn diagram indicating the number of common and uniquely identified differentially regulated genes between the Hfq input and co-IP data sets. Numbers represent genes identified by DESeq2 as significantly differentially expressed with a log2 fold-change in expression of at least 1.5 when comparing the WT (KR10000) and Δpnp (NRD999) strains. (B) RNA-seq read coverage of short RNA library preparations of input RNA and RNA co-immunoprecipitated with Hfq for the WT (KR10000; black) and Δpnp (NRD999; red) strains for the same regions shown in Figure 1B. Coverage represents depth per million paired-end fragments and was averaged between two normalized replicates. (C) Input and Hfq co-immunoprecipitation fractions probed by northern blot for the presence of the RNA fragments shown in B in the wild-type (KR10000) and Δpnp (NRD999) strains. SsrA served as a loading control. An expanded view of the northern blots is shown in Supplementary Figure S2.