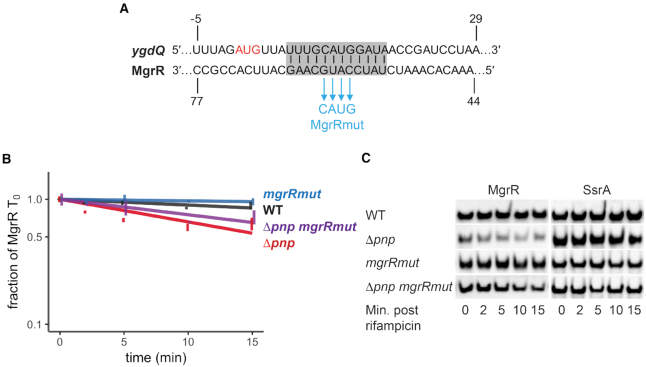
Figure 6.
MgrR target-pairing mutations partially reduce its decay that occurs in the absence of PNPase. (A) Illustration showing complementarity between MgrR and ygdQ (highlighted in grey) and four nucleotide changes (blue) that were introduced into mgrR (MgrRmut). The start codon of ygdQ is depicted in red. (B) Stability curves of the MgrR mutant in WT and Δpnp backgrounds. Overnight cultures of strain NRD1138 (WT rph-1) and derived strains carrying the pnp deletion (Δpnp, NRD1139) and harboring mutations in mgrR (mgrRmut, NRD1599; Δpnp mgrRmut, NRD1601) were diluted into fresh LB medium and grown to early exponential phase (OD600 of 0.3). Rifampicin RNA stability time courses were performed over 15 min and MgrR was examined by northern blot as described in the Materials and Methods using the probe MgrR2. MgrR levels were normalized to the control RNA SsrA and graphed as a fraction of initial MgrR level (T0). Results represent the mean of three independent experiments and the error bars represent the standard error of the mean. (C) Representative blots for MgrR and SsrA.