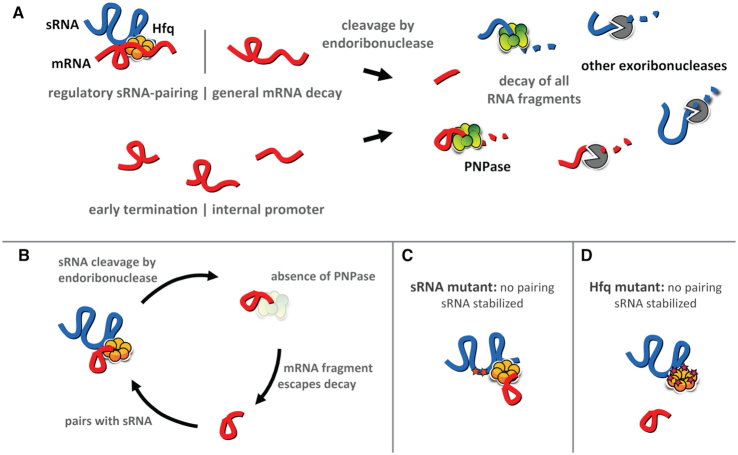
Figure 8.
A model for the role of PNPase in sRNA-mediated gene regulation. (A) mRNA fragments are first produced by endoribonuclease cleavage due to regulatory sRNA pairing or general mRNA decay, or directly via early transcription termination or by internal promoters. In the presence of PNPase, mRNA fragments are completely digested. (B) In the absence of PNPase, some mRNA fragments escape decay by other exoribonucleases. Those that retain Hfq binding and sRNA pairing sequences may interact with sRNAs and promote their cleavage by endoribonucleases. (C, D) In the absence of PNPase, sRNA stability may be restored by disrupting sRNA–mRNA pairing, either by mutating the pairing region of the sRNA (C) or by mutating the corresponding mRNA-binding site of Hfq (D).