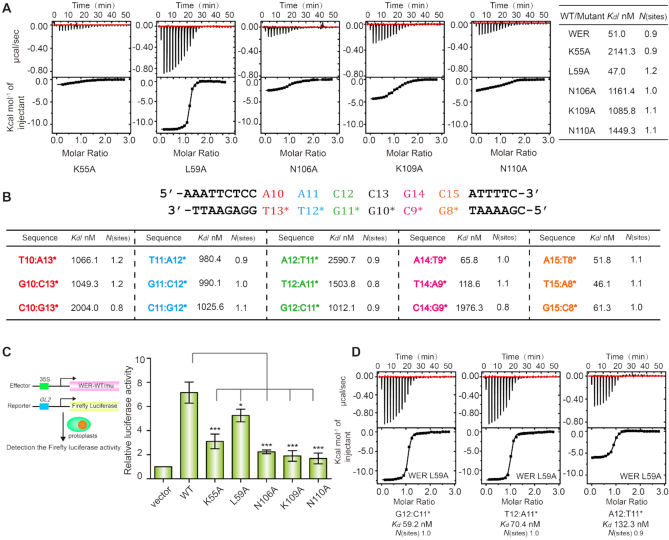
Figure 3.
Verification of sequence-specific interactions. (A) ITC experiments showing the impact of mutation of WER residues involved in dsDNA (5′-AAATTCTCCAACCGCATTTTC-3′, 5′-GAAAATGCGGTTGGAGAATTT-3′) binding. (B) ITC experiments showing the impact of DNA core motif (5′-AACCGC-3′) mutation on WER binding. The detailed sequence of the target DNA is shown on the top of the table. For mutated DNAs, only the mutated base pairs are listed in the table for clarity. (C) Dual luciferase assay of the GL2 promoter activation activity of wild-type and mutated WER proteins. Values are means ± SD of three independent biological replicates. * and *** indicate statistically significant differences between the wild type (WT) and mutant at P < 0.05 and P < 0.001, respectively. (D) ITC experiments showing the binding affinities of WER L59A mutant to the A12:T11*, G12:C11* and T12:A11* mutants of the 5′-AACCGC-3′ motif, respectively.