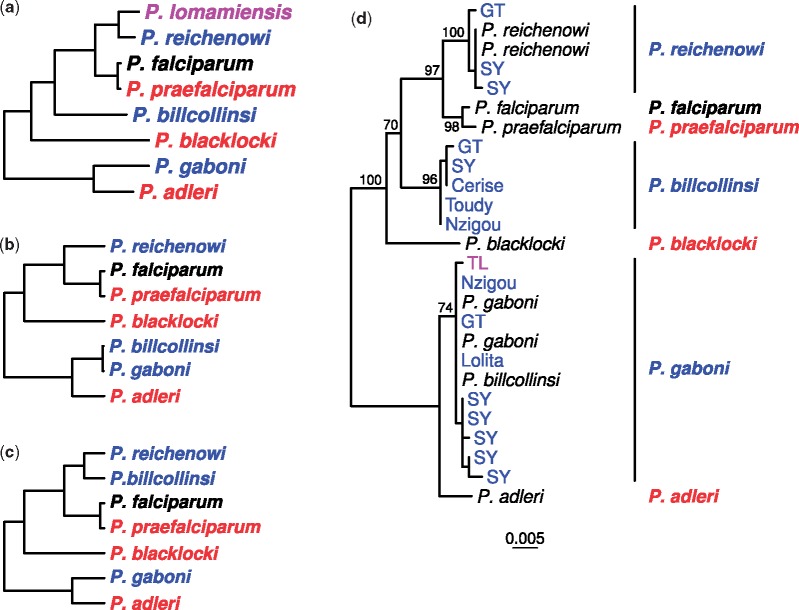
Fig. 1.
—Relationships among Laverania species. (a) The standard topology for the eight known Laverania species (Liu et al. 2010, 2016, 2017). Colors indicate the parasite’s usual host species: Bonobos (purple), chimpanzees (blue), humans (black), and gorillas (red). Although most prevalent in chimpanzees, P. gaboni (and on rare occasions P. reichenowi) also infects bonobos (Liu et al. 2017). The tree was derived by maximum likelihood analysis of a 3.4 kb region of the mitochondrial genome (see Supplementary Fig. 1a in Liu et al. 2010). (b) Schematic representation of an anomalous topology with transfer from P. gaboni to P. billcollinsi (termed “topology C” in Supplementary Figure 5 of Otto et al. 2018). (c) Schematic representation of an anomalous topology with transfer between P. reichenowi and P. billcollinsi (termed “topology D” in Supplementary Figure 5 of Otto et al. 2018). (d) Maximum likelihood tree of one of four genes (gene ID PF3D7_1328000) hypothesized to have resulted from P. gaboni to P. billcollinsi gene transfer (Otto et al. 2018). Sequences were obtained from published reference genomes (black), limiting dilution PCR from ape blood or fecal samples collected at three field sites (SY, GT, TL), or de novo assembled contigs of sequencing data released by Otto et al. (labelled with the name of the animal from which the data were derived: Toudy, Nzigou, Cerise, Lolita; see Supplementary Table 1 of Otto et al. 2018). For PCR-derived sequences and new contigs, color indicates whether the sample was obtained from a chimpanzee (blue) or a bonobo (purple). Note that the sequence from the published P. billcollinsi reference genome clusters within P. gaboni, whereas other P. billcollinsi sequences fall in the expected position within the tree. The scale bar represents 0.005 nucleotide substitutions per site; bootstrap values from 100 replicates are shown for interspecific branches with at least 70% support.