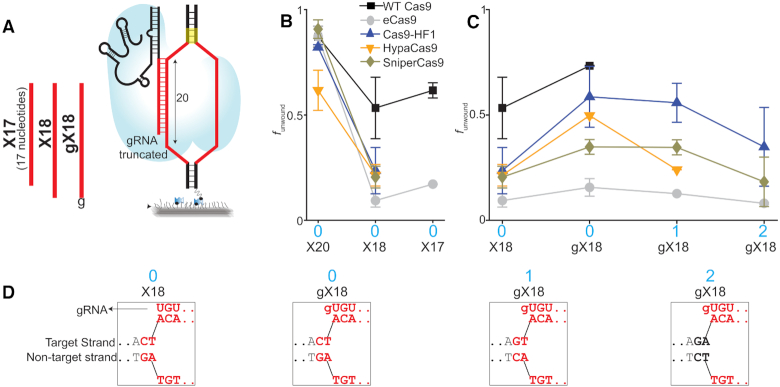
Figure 4.
Unwinding by truncated gRNA. (A) Schematic and naming convention of truncated gRNA used in the unwinding assay. X18/X17 indicate the gRNA with length of 18/17 base pairs in the target strand hybridizing region, as opposed to the length of 20 bp for canonical gRNA. Addition of a mismatched g at the 5′ end of X18 gives gX18. (B) Comparison of funwound between X20, X18, and X17 for cognate DNA. (C) funwound with the different gRNA–Cas9 combinations for different DNA with 18 base pairs between gRNA and target strand. (D) The PAM-distal sequences of such gRNA and DNA combination are shown below for each column. All these DNA targets present the same number of 18 matching sequences to the gRNAs but differ in the sequence only beyond these 18 matching sequences at the PAM-distal site. Such differing nucleotides are marked in black. Data for dCas9, deCas9 and dCas9–HF1 with X20 is taken from a previous study (31). Error bars represent SD.