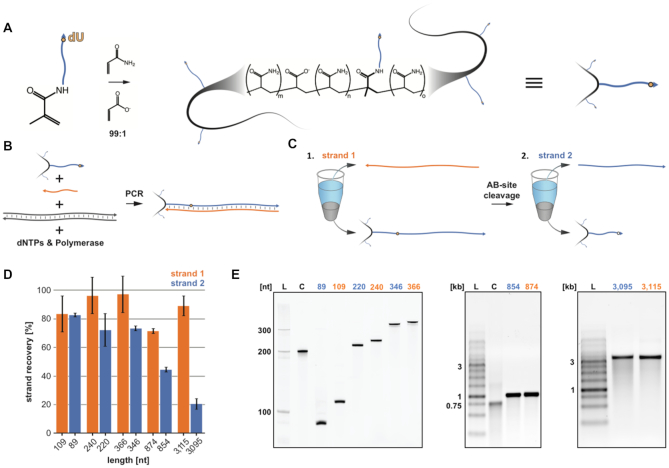
Figure 1.
MeRPy-PCR overview and recovery yields for strands 1 (untagged) and 2 (initially tagged) of different amplicon lengths. (A) Production of the polymer-tagged primer. A 5′ acrydite modified primer is polymerized with acrylamide and sodium acrylate (ratio 99:1) to form a long linear DNA-tagged polymer. (B) MeRPy-PCR procedure following standard PCR guidelines. (C) (1.) Recovery of strand 1 under alkaline denaturing conditions and methanol precipitation. (2.) Recovery of strand 2, after treatment with UDG and DMEDA followed by a methanol precipitation. (D) Recovery yield for strand 1 and 2 of various lengths. Bar graphs denoting the recovery yield (%). Strand recovery yield was determined by the absolute recovered strand yield (pmol) relative to MeRPy-PCR input (pmol). Data are shown as mean ± STD (N = 3). (E) Gel electrophoresis of MeRPy-PCR derived ssDNA. Left, denaturing polyacrylamide gel with L – 20 bp Ladder, C – 200mer control from Integrated DNA Technologies (IDT). Middle and right, native agarose gels with L – 1 kb Ladder, C–750mer control from IDT. MeRPy-PCR derived and commercial ssDNAs were loaded with normalized mass amounts for each gel lane in (E).