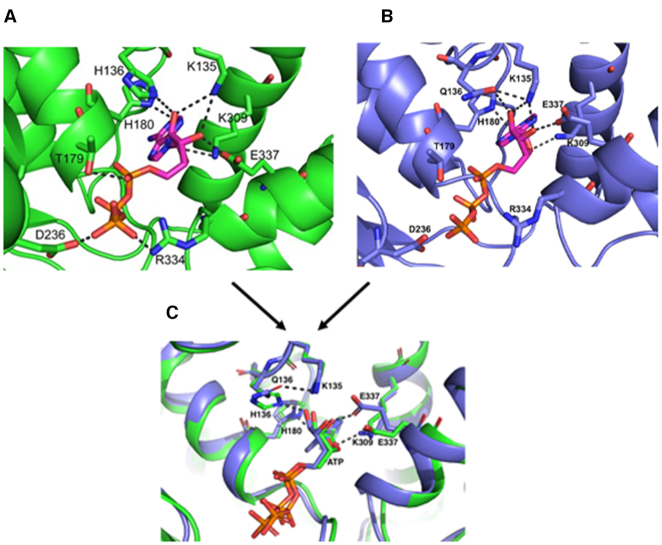
Figure 6.
His136 residue of DnaA participates in ATP binding. Homology model of E. coli DnaA (amino acid residues 130–369) was constructed upon an A. aeolicus DnaA structure (PDB ID: 1L8Q) using the SWISS-MODEL server. The crystal structure of A. aeolicus DnaA bound to AMPPCP (PDB ID: 2HCB) was used to perform docking of ATP into the active site of E. coli DnaA and the final model was subsequently energy minimized using Amber software. The ATP binding pocket in DnaA WT (A) and in DnaA Gln136 mutant (B). The structures in (A) and (B) are superimposed in (C).