Table 1.
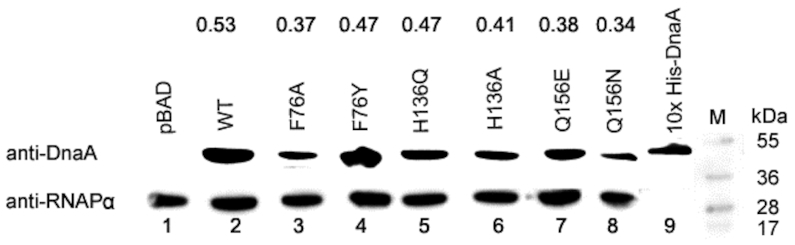
Substitutions at H136 does not support initiation of replication in vivo: E. coli EH3827 cells were transformed with pZL606 and pRS9-pRS14 plasmids and the transformants were grown for 10–12 generations. Proteins were expressed by adding 0.2% arabinose and growing the cultures for an additional 90 min. Protein lysates were prepared and the protein amounts in the lysate were estimated using the Bradford assay by loading equal amounts of total protein in each lane (lanes 1–9). Samples were analyzed by SDS-PAGE and after transfer to the PVDF membranes and western blotting. The blots were probed with polyclonal anti-DNA antibody and reprobed with anti-RNAP antibody (treated as a loading control). A representative scan is presented here to show the expression level of DnaA. Numbers indicate the relatives ratios of DnaA protein to total protein in each lane. Next, cells containing different dnaA alleles under arabinose-inducible promoter were made competent and transformed with plasmid containing oriC. The transformed cells were plated at LB-Agar plates supplemented with 0.2% arabinose and appropriate antibiotics (see material and methods). Data represent average values from three biological replicates
| ||
|---|---|---|
| E. coli EH3827 | Colony forming unit (cfu)/μg | |
| With | Ampicillin (0.2% arabinose) |
Ampicillin + chloramphenicol (0.2% arabinose) |
| pBAD | 1.9 × 107 | <1 |
| pZL606(dnaA) | 2.0 × 107 | 2.5 × 104 |
| pRS9(dnaAF76Y) | 4.4 × 107 | 1.6 × 104 |
| pRS10(dnaAF76A) | 2.8 × 107 | 3.4 × 104 |
| pRS11(dnaAH136Q) | 3.4 × 107 | <1 |
| pRS12(dnaAH136A) | 3.0 × 107 | <1 |
| pRS13(dnaAQ156E) | 2.4 × 107 | 3.4 × 104 |
| pRS14(dnaAQ156N) | 2.5 × 107 | 3.4 × 104 |
