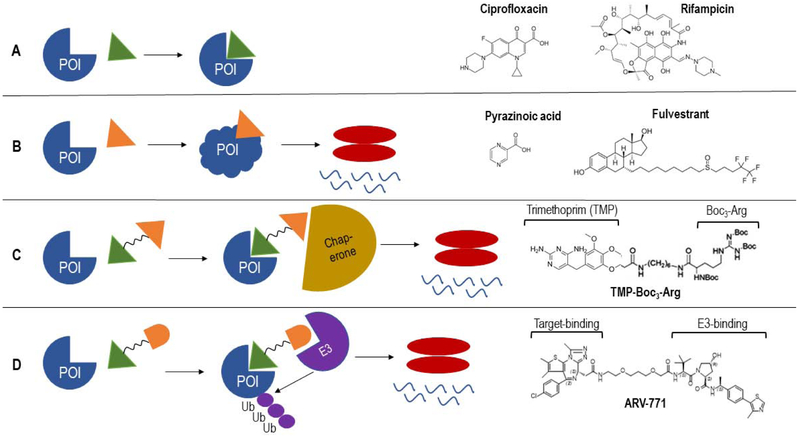
Fig. 1.
Examples of drugs either interfering with the activity of their target (A) or triggering degradation of their respective targets by various mechanisms (B,C,D). (A) Drug binds its specific target and alters target activity as a result of this interaction. Examples of such antibiotics are Ciprofloxacin and Rifampicin, which bind and inhibit the bacterial DNA gyrase and RNA polymerase respectively. (B) Drug binding to its target causes conformational changes of target, that results in target degradation. Pyrazinoic acid (POA) binds to its target Mycobacterium tuberculosis aspartate decarboxylase which changes its conformation and undergoes degradation via the Mtb ClpC1-P1P2 machinery (Gopal et al., 2019). Fulvestrant triggers degradation of its target Estrogen receptor ERα via the mammalian proteasome by changing its surface hydrophobicity (Wu et al., 2005). (C) Drug binds target, hydrophobic tag recruits chaperone protein which unfolds and transports the target to the cell’s degradation machinery. Example is Trimethoprim-Boc3-Arg, where Trimethoprim binds to E. coli DHFR (when expressed in mammalian cells) and the hydrophobic Boc3-Arg moiety tags it for degradation (Long et al., 2012). (D) PROTACS: bifunctional molecules with one end of the drug binding the target and the other end binding an E3 ligase which ubiquitinates target and recruits the proteasome to degrade the target. Example is ARV-771 which binds to Bromodomain and extra-terminal (BET) protein via the target-binding moiety and the E3 ligase Cereblon via the E3-binding moiety, thus resulting in degradation of BET proteins (Raina et al., 2016). POI, protein of interest.