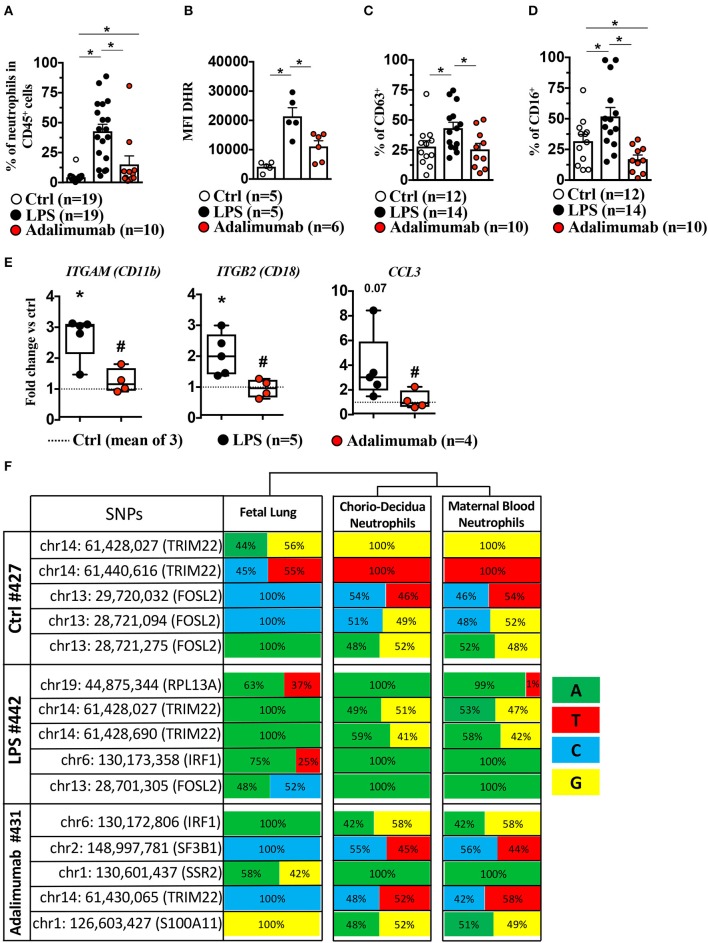
Figure 4.
Adalimumab decreased LPS-induced chorio-decidua neutrophil accumulation and activation. Chorio-decidua cells were scraped and digested with protease/DNAase, and single cell suspensions were used for multiparameter flow cytometry. Neutrophils were defined as CD45+CD3−CD14lowHLADR-CD88+ CD56− cells. Inhibition of TNF-signaling by Adalimumab decreased significantly neutrophil frequency (Ctrl n = 19; LPS n = 19; Adalimumab n = 10) (A), neutrophil reactive oxygen species production measured by Dihydro-rhodamine (DHR) fluorescence (Ctrl n = 5; LPS n = 5; Adalimumab n = 6) (B), CD63 expression, a marker of degranulation of azurophilic granules (Ctrl n = 12; LPS n = 14; Adalimumab n = 10) (C), and expression of FCγIII receptor CD16 (Ctrl n = 12; LPS n = 14; Adalimumab n = 10) (D). Data are mean ± SEM, *p < 0.05 between comparators by Mann-Whitney U-test. (E) Adalimumab decreased significantly the expression of genes associated with neutrophil chemotaxis. The box plot shows fold-change of gene expression in chorio-decidua neutrophils normalized to control chorio-decidua neutrophils (dotted-line represents the mean of 3 Ctrl; LPS n = 5; Adalimumab n = 4). *p < 0.05 vs. ctrl; #p < 0.05 vs. LPS (Mann–Whitney U-test). (F) Frequency of nucleotides in SNPs in the fetal lung (representing fetal genome), chorio-decidua neutrophils, and maternal blood neutrophils (representing maternal genome) at five different informative loci in each mother-infant pair (total n = 3). Note similar frequency distribution of nucleotides at each of the SNPs in maternal blood neutrophils and chorio-decidua neutrophils and divergence compared to the fetal lung. For each SNP the chromosomal location and the encoded gene is shown. The frequency of nucleotides was computed at each SNP based on the number of reads shown in Supplementary Table 6.