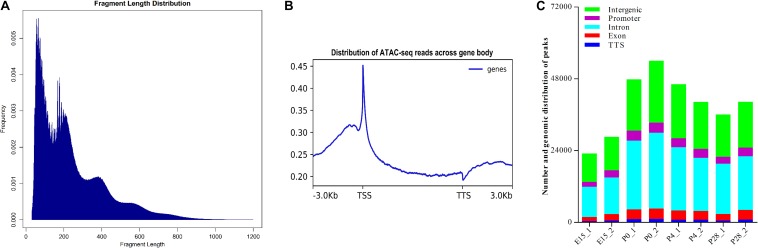
FIGURE 4.
Quality estimation, peak calling, and genomic distribution of ATAC-seq reads during early ovarian development. (A) Frequency distribution of fragment lengths within a representative ATAC-seq library. The smallest fragment peaks represent sequencing reads in inter-nucleosome open chromatin, while larger peaks represent those spanning nucleosomes. (B) Distribution plot of sequencing reads from a representative ATAC-seq library across all genes. We normalized all genes according to their lengths and calculated the average ATAC-seq signals between TSS (–3 kb) and TTS (+ 3 kb) of all genes after peak calling. (C) Number and genomic distribution of peaks identified by ATAC-seq in each sample. Genomic annotations included the promoter (± 1 kb of TSS), TTS (between –100 and + 1 kb of TTS), exon, intron, and intergenic region, and the front region was regarded as the final annotation according to the above order if overlap occurred.