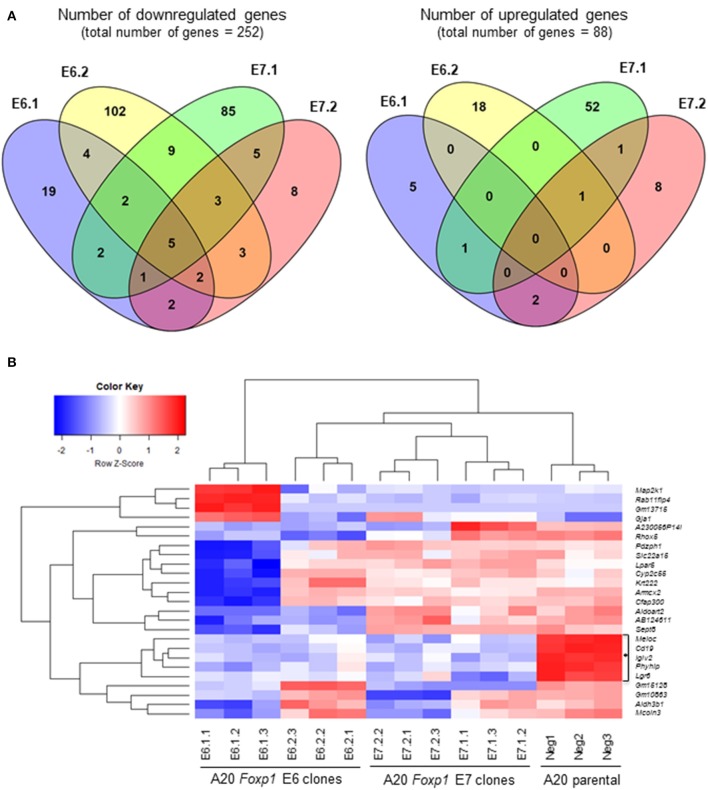
Figure 4.
Foxp1 depletion in CRISPR/Cas9 clones alters gene expression and signatures involved in human DLBCL pathogenesis. (A) Analysis of differential gene expression was performed after RNA sequencing, comparing the gene expression profile of each clone with that of the A20 parental cell line. Venn diagrams show numbers of genes down or upregulated following Foxp1 knockout (two-fold cut off, FDR adjusted p-value, p < 0.05). The number of individual genes downregulated in each clone is E6.1 (n = 37), E6.2 (n = 130), E7.1 (n = 112), E7.2 (n = 29). The number of individual genes upregulated in each clone is E6.1 (n = 8), E6.2 (n = 19), E7.1 (n = 55), E7.2 (n = 12). Gene lists produced from this analysis are provided in Supplementary Table 7. (B) Heat map representation of top 25 significantly (p < 0.05) differentially expressed genes in Foxp1 CRISPR/Cas9 edited clones compared to A20 parental cells. The genes commonly downregulated in all the Foxp1 CRISPR clones are indicated by a bracket and asterisk. Unsupervised clustering by complete-linkage Euclidean distance was used to generate the heat map dendrograms.