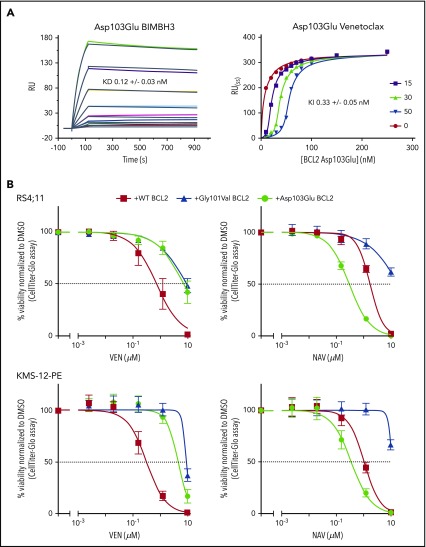
Figure 2.
Venetoclax/BIM binding characteristics and in vitro sensitivity of BCL2 Asp103Glu mutation. (A) Impact of Asp103Glu on the ability of BCL2 to bind BH3 ligands. (Left) BIMBH3 binding. A total of 0 to 40 nmol/L mutant BCL2 was used as an analyte against the BIMBH3 peptide immobilized on a surface plasmon resonance (BIAcore) sensor chip. The raw response (RU) curves (colored curves) from a representative experiment were fitted to 1 site-specific kinetic model (black curves) to derive on and off rates (supplemental Table 2), and calculate KD values for interactions with Asp103Glu. (Right) Steady-state competition of various venetoclax concentrations (0-50 nmol/L) prebound to Asp103Glu (0-250 nmol/L), competing against a BIMBH3 immobilized chip. Fitted data were used to derive the KI for venetoclax binding to Asp103Glu. Data are representative of 3 independent experiments, reporting means ± 1 SD. (B) Expression of BCL2 Asp103Glu in RS4;11 (top) or KMS-12-PE (bottom) cell lines reduces sensitivity to venetoclax (left) but not to navitoclax (right). Each of these mutants or wild-type (WT) BCL2 were expressed and the in vitro sensitivities to venetoclax (0-10 μmol/L) or to navitoclax (NAV; 0-10 μmol/L) measured 24 hours later. Data represent means ± 1 SD of at least 3 independent experiments. KD,equilibrium binding constant for BIMBH3 peptide, from direct binding experiments; KI, fitted equilibrium binding constant for venetoclax, from steady-state competition experiments.