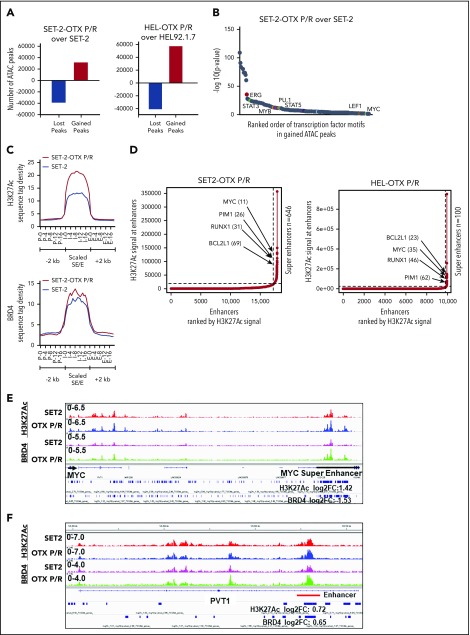
Figure 2.
Analysis of the epigenome in OTX-P/R sAML cells demonstrates greater presence of H3K27Ac and increased BRD4 occupancy. (A) Number of gained and lost peaks in SET-2-OTX P/R and HEL-OTX P/R cells compared with parental SET-2 and HEL cells as determined by ATAC-seq analysis. (B) TF binding motif analysis was conducted utilizing HOMER. The −log10 P values of the rank-sorted motifs in the gained ATAC-seq peaks of SET-2-OTX P/R cells compared with parental SET-2 cells are shown. (C) Sequence tag density of H3K27Ac and BRD4 within Es and SEs vs 2 kb upstream or downstream in SET-2-OTX P/R compared with SET-2 cells. (D) Rank ordering of SEs (ROSE) analysis was performed on the H3K27Ac ChIP-seq peaks from SET-2-OTX P/R and HEL-OTX P/R cells. The numbers indicate the rank of the SE out of the total number of identified SEs in each P/R cell line. (E-F) Integrated Genomics Viewer plots of H3K27Ac and BRD4 signal densities at the MYC SE and PVT1 gene in SET-2 and SET-2-OTX P/R cells. The log2 fold change (log2FC) in peak numbers for H3K27Ac and BRD4 was calculated with diffReps. The fold changes for significant alterations (P < .05) are noted beneath the SEs/Es.