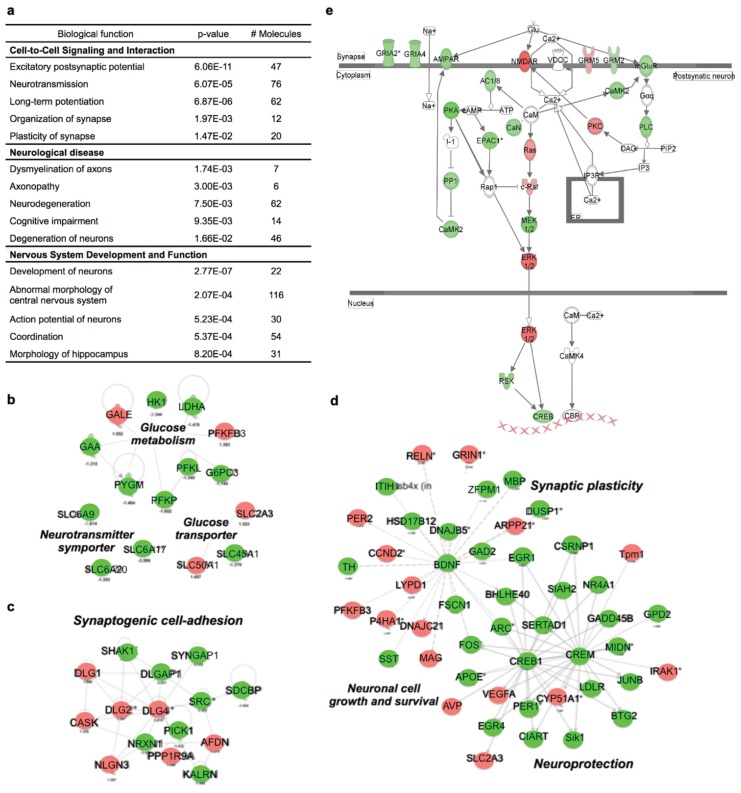
Figure 2.
Dehydration alters hippocampal transcriptional networks. A hippocampal transcriptional network analysis was performed, comprising next-generation RNA sequencing and bioinformatic analyses. (a) Genes that were differentially regulated between the control (CON) and dehydrated (DEH) mice were classified into functional categories via an Ingenuity Pathway Analysis (IPA). Significantly changed biological functions are indicated by p-values calculated using a Fisher’s exact test, and the number of distinctively modulated molecules in each functional category is indicated. (b–d) Transcriptional networks related to altered glucose metabolism, synaptogenic cell-adhesion, and synaptic plasticity were differentially regulated in response to long-term dehydration. (e) Genes associated with long-term potentiation (LTP) signaling transduction were differentially up- (red) or downregulated (green) in the DEH compared to the CON mice.