Figure 2.
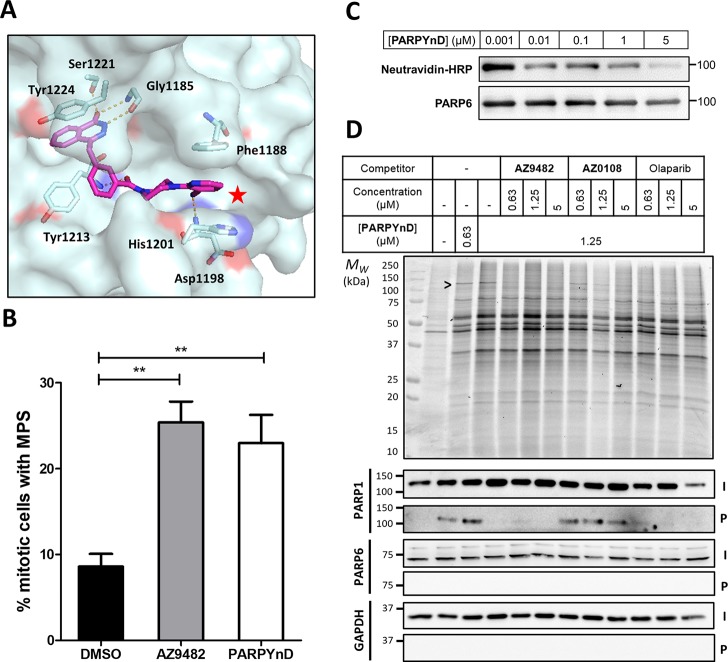
Validation of PARPYnD as a suitable probe for parent molecule profiling. (A) Crystal structure of parent molecule AZ9482 bound in the NAD+-binding pocket of PARP5a (PDB ID: 5ECE), with key interactions highlighted in orange. The red star highlights the solvent exposed position at which modification is expected to minimally perturb the binding of a probe into this pocket of the PARP enzymes. (B) Quantification of the percentage of mitotic cells with MPS phenotype (>2 spindle poles per cell) after treatment with AZ9482 (N = 2) and PARPYnD (N = 2), both at 41 nM, versus DMSO (N = 4); double asterisk (**) represents raw P value <0.001 in unpaired Student’s t test; raw data found in the Supporting Information, Extended Data S1. (C) PARP6 activity assay: recombinant GST-tagged PARP6 was incubated with biotinylated NAD+ and varying concentrations of PARPYnD. GST-PARP6 auto-MARylation was measured by immunoblotting against NeutrAvidin-HRP; decreased signal with increasing PARPYnD concentration indicated catalytic inhibition of PARP6. (D) Gel and Western blot analysis of live cells labeled with PARPYnD and ligated to AzTB (Figure S3A) with/without cotreatment with various competitor molecules. In-gel fluorescence was used to qualitatively assess TAMRA-tagged proteins, and streptavidin-based enrichment and immunoblot analysis was used to validate specific targets. I = input (whole lysate); P = pull down (enriched fraction); > indicates competition seen on gel, validated as PARP1 by immunoblot. Uncropped gels and immunoblots associated with all figures can be found in Figure S7.