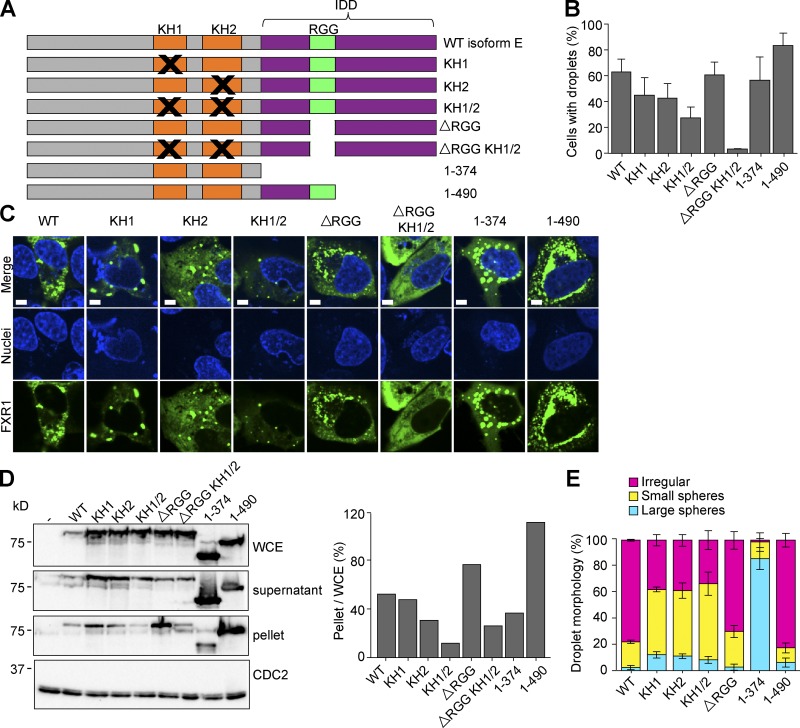
Figure 7.
Phase separation is RNA binding dependent. (A) Location of mutations in the domains of isoform E. (B and C) U2OSΔFFF cells were transfected with WT PCMV2xTetO2-mNeonGreen-FXR1 isoform E (pAGB1162), plasmids with mutated KH1 (pAGB1171), KH2 (pAGB1172), KH1/2 (pAGB1173), RGG (pAGB1174), RGG and KH1/2 (pAGB1175), complete IDD (pAGB1176), or partial IDD (pAGB1177). All constructs were induced with 25 ng/ml Tet to endogenous levels. Black Xs indicate mutated regions. (B) Quantification of droplet formation. n > 350 cells from three experiments. (C) Representative images of cells expressing the different mutants. Scale bar, 5 µm. (D) Whole-cell extracts (WCE), lysate, and pellets were analyzed by Western blotting using an antibody against FXR1. Right: percent protein in the pellet quantified as in Fig. 5. (E) Quantification of droplet morphology using the same three categories that were used in Fig. 5. Data are means ± SEM.