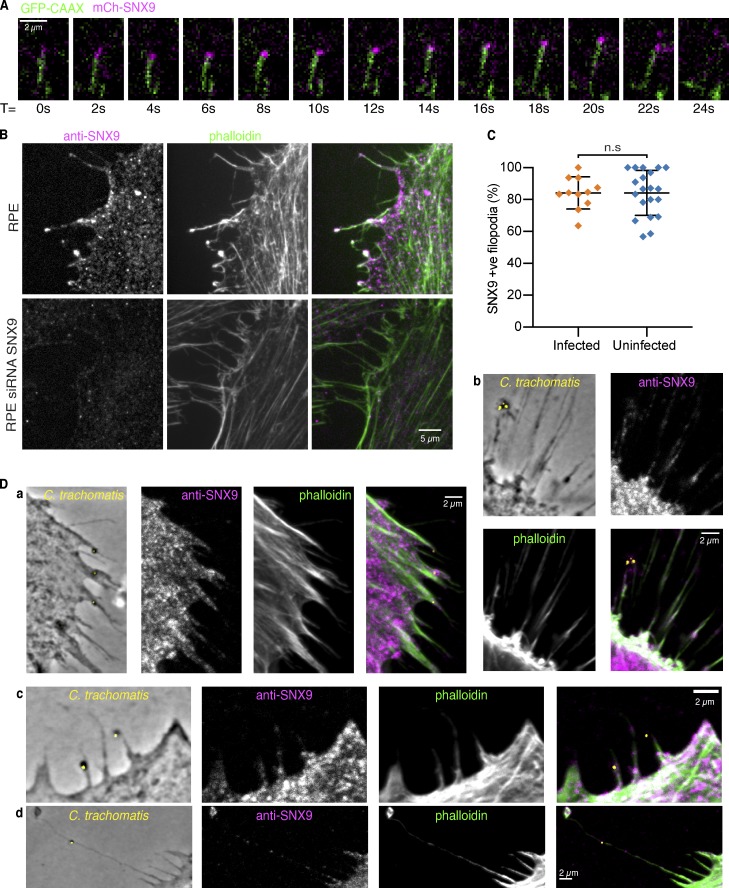
Figure 4.
SNX9 localizes to filopodia in Xenopus gastrula explants and human cultured cells. (A) Background-subtracted single frames from a TIRFM time-lapse sequence taken at 2-s intervals of Xenopus chordamesoderm cells expressing mCh-SNX9 and GFP-CAAX membrane marker shows SNX9 tracking a growing filopodium tip. Scale bar, 2 µm. (B) Confocal images of RPE-1 cells treated with either scramble or SNX9 siRNA and immunolabeled with rabbit anti-human SNX9 and phalloidin; endogenous SNX9 is in RPE-1 cell filopodia and is abolished on knockdown. Scale bar, 5 µm. (C) Quantification of SNX9 in filopodia in uninfected RPE-1 and in cells infected with C. trachomatis. Each data point represents an individual imaging region; lines indicate mean ± SD. For infected cells, n = 248 filopodia from 62 cells across n = 11 images, for uninfected, n = 416 filopodia from 139 cells across n = 20 images. Unpaired t test, P = 0.9958 (n.s.). (D) a–d: Example images of phase contrast and fluorescence confocal images of RPE-1 cells infected with C. trachomatis and immunolabeled for SNX9 and phalloidin show C. trachomatis sticking to filopodia containing SNX9. Scale bars, 2 µm.