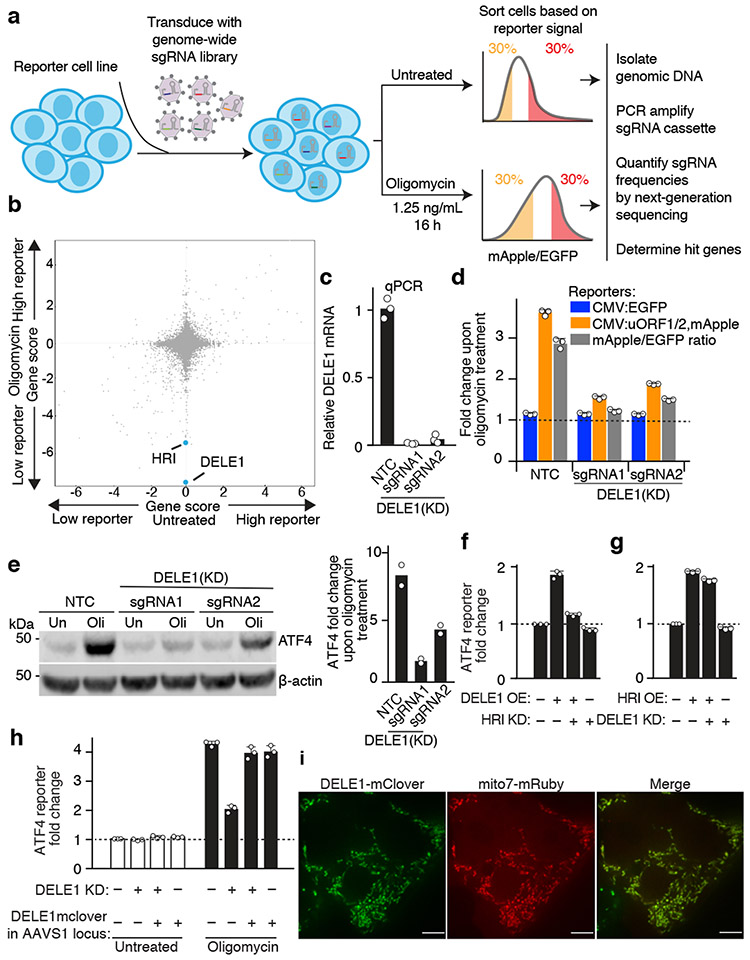
Fig. 2. A CRISPRi screen identifies a requirement for DELE1 in ATF4 induction.
(a) Schematic of screen.
(b) Comparison of gene scores (defined in Methods) from the screen in untreated and oligomycin-treated conditions.
(c) Knockdown of DELE1 quantified by qPCR (n = 3 technical replicates).
(d) Reporter activation in cells expressing a non-targeting control sgRNA (NTC) or DELE1 sgRNAs (mean ± s.d., n = 3 culture wells).
(e) Immunoblot of endogenous ATF4. Cells were treated with 1.25 ng/mL of oligomycin for 16 hrs (Oli) or untreated (Un). Left, representative blot; Right, quantification of n = 2 blots.
(f) ATF4 reporter activation in cells transiently overexpressing (OE) DELE1-mClover and expressing sgRNA to knockdown (KD) HRI where indicated. (mean ± s.d., n = 3 culture wells).
(g) ATF4 reporter activation in cells transiently overexpressing (OE) HRI and expressing sgRNA to knockdown (KD) DELE1 where indicated. (mean ± s.d., n = 3 culture wells).
(h) DELE1-mClover expression from the AAVS1 safe-harbor locus is not sufficient to induce the ATF4 reporter in the absence of stress, but complements DELE1 knockdown upon oligomycin treatment (mean ± s.d., n = 3 culture wells).
(i) Co-localization of DELE1-mClover with the mitochondrial-targeted mRuby (Mito7-mRuby). Scale bar, 7 μm. Similar results for n = 2 culture wells.