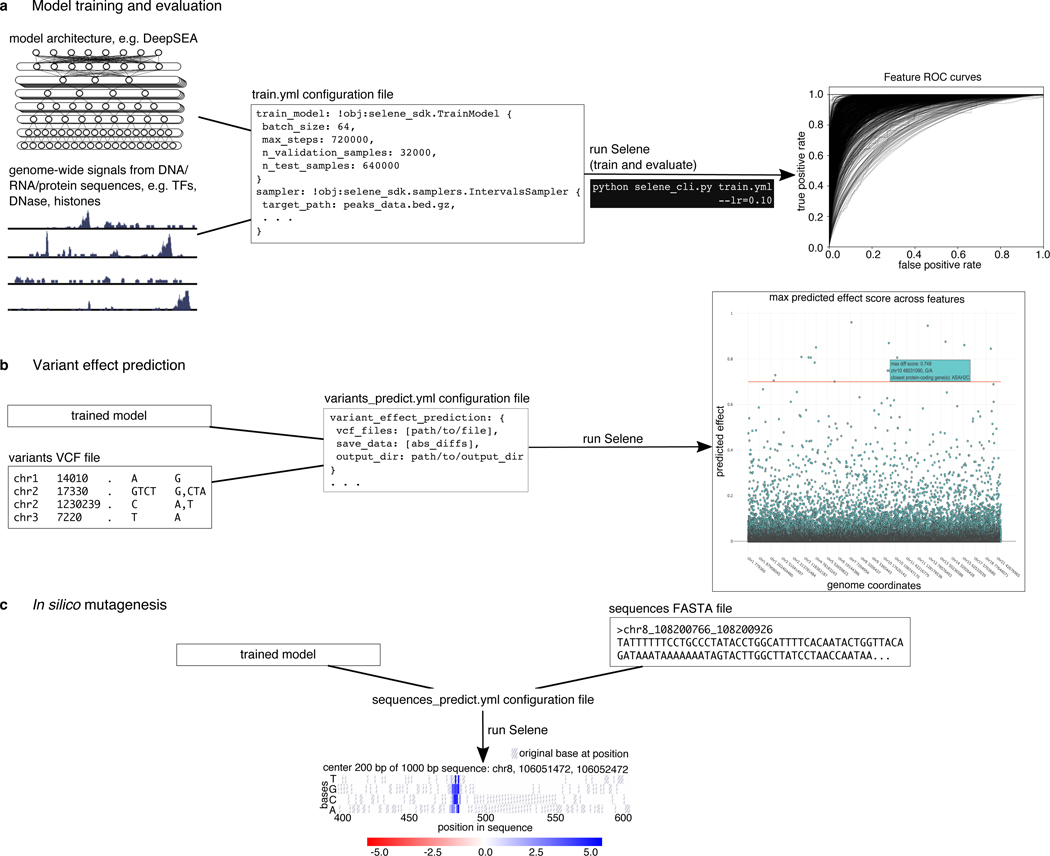
Figure 1. Overview of Selene.
(a) Selene enables users to train and evaluate new deep learning models with very few lines of code. As input, the library accepts (left) the model architecture, dataset, and (mid) a configuration file that specifies the necessary input data paths and training parameters; Selene automatically splits the data into training and validation/testing, trains the model, evaluates it, and (right) generates figures from the results. (b) Selene also supports the use of trained models to interpret variants. In addition to being able to run variant effect prediction with the same configuration file format, Selene includes a visualization of the variants and their difference scores as a Manhattan plot, where a user can hover over each point to see variant information. (c) Users interested in finding informative bases that contribute to the binding of a certain protein can run Selene to get mutation effect scores and visualize the scores as a heatmap.