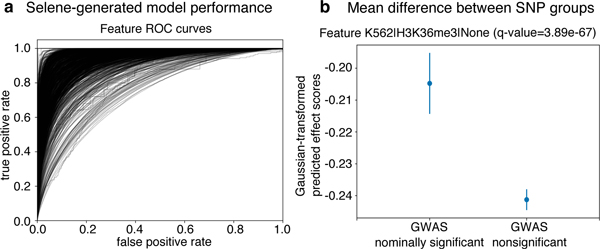
Figure 3. Using Selene to train a model and obtain model predictions for variants in an Alzheimer’s GWAS study.
(a) Selene visualization of the performance of the trained model from case-study 2. (b) The trained model, on average, predicts greater differences for the nominally significant variants (p-value < 0.05, n = 422398) reported in the IGAP early onset Alzheimer’s GWAS study16 compared to those that are nonsignificant (p-value > 0.50, n = 3842725). Here we visualize the mean and 95% confidence intervals of the quantile-normalized (against the Gaussian distribution) predicted effect scores of the two variant groups for the genomic feature H3K36me3 in K562 cells, the feature in the model with the most significant difference (one-sided Wilcoxon rank-sum test, adjusted p-value using Benjamini—Hochberg of 3.89 ✕ 10−67). After applying the multiple testing correction, 914 of the 919 features that the model predicts showed a significant difference (ɑ < 0.05) between the groups.