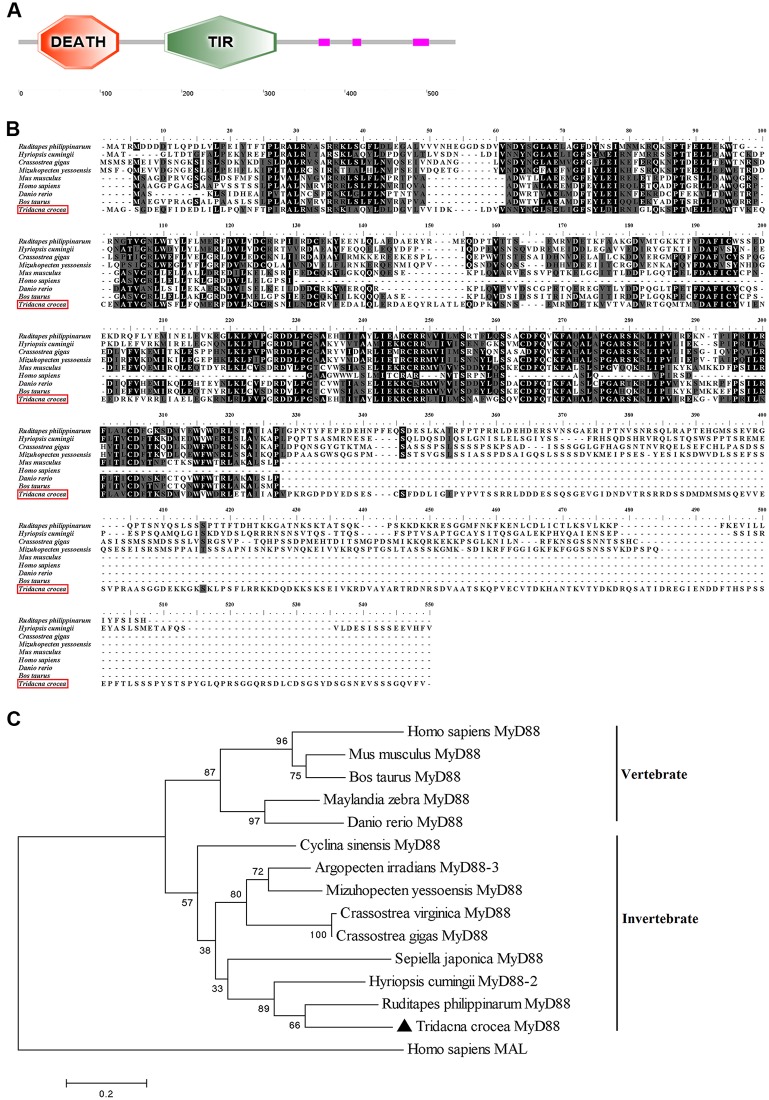
Fig 4. Sequence analysis of TcMyD88.
(A) Functional domain organization of TcMyD88. (B) Multiple sequence alignment analysis of TcMyD88 and other known members of the MyD88 family. The black shaded sequences indicate residues that exactly match the consensus. The gray shaded sequences showed a similar relationship. The GenBank accession numbers corresponding to the MyD88 sequences examined are listed: Ruditapes philippinarum (AEF32114.1), Hyriopsis cumingii (AHB62785.1), Crassostrea gigas (NP_001292286.1), Mizuhopecten yessoensis (XP_021355224.1), Mus musculus (NP_034981.1), Homo sapiens (NP_001166037.2), Danio rerio (NP_997979.2), and Bos taurus (NP_001014404.1). (C) A phylogenetic tree showing the relationships among TcMyD88 and MyD88 from different species was constructed by the neighbor-joining method using MEGA5.0 software. The node values represent the percent bootstrap confidence derived from 1000 replicates. The GenBank accession numbers corresponding to the MyD88 sequences and MAL sequence examined: Argopecten irradians (AVP74319.1), Crassostrea virginica (XP_022332088.1), Cyclina sinensis (AIZ97751.1), Sepiella japonica (AQY56781.1), Maylandia zebra (XP_004546813.1), Homo sapiens MAL (NP_002362.1), and other sequences referenced in Fig 4B.