FIG 2.
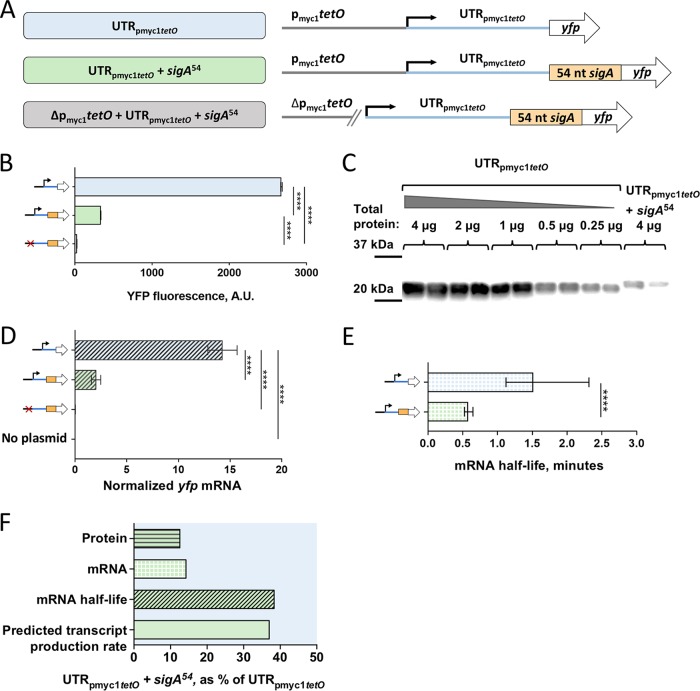
The first 54 nt of the sigA coding sequence affects transcript production rate and mRNA half-life. (A) Constructs transformed into M. smegmatis to determine the impact of the first 54 nt of the sigA coding sequence (sigA54) on expression of a YFP reporter. (B) Median YFP fluorescence of strains bearing the constructs in panel A, determined by flow cytometry. Error bars denote 95% CI. Strains were compared by Kruskal-Wallis test followed by Dunn’s multiple-comparison test. (C) Lysates from strains bearing constructs with and without sigA54 were subject to Western blotting to detect the C-terminal 6×His tag on the YFP. The mass of total protein loaded per lane is stated. (D) yfp mRNA abundance for strains bearing the indicated constructs, determined by qPCR and normalized to expression of endogenous sigA. Error bars denote standard deviation. Strains were compared by analysis of variance (ANOVA) with Tukey’s honestly significant difference (HSD) test. (E) The half-lives of yfp mRNA produced from the indicated constructs were measured. Error bars denote 95% CI. Half-lives were compared using linear regression analysis (n = 3). (F) Protein abundance, mRNA abundance, mRNA half-life, and calculated transcript production rate for the construct containing sigA54 are shown as a percentage of the values produced by a construct that lacks sigA54 but is otherwise identical. ****, P < 0.0001.