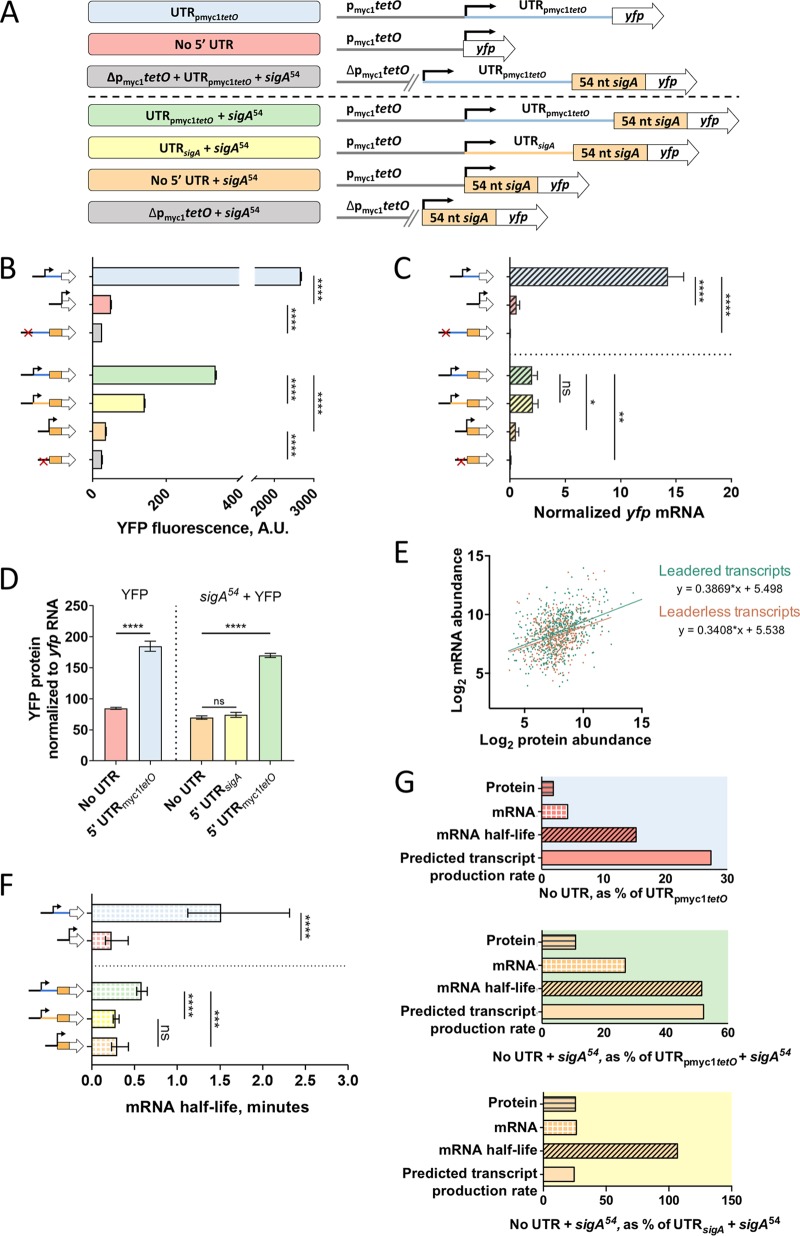
FIG 4.
Leaderless transcripts have altered translation efficiencies, mRNA half-lives, and predicted transcript production rates compared to those of leadered controls. (A) Constructs transformed into M. smegmatis to compare leaderless versus leadered gene structures. (B) Median YFP fluorescence of strains bearing the constructs in panel A, determined by flow cytometry. Error bars denote 95% CI. Strains were compared by Kruskal-Wallis test followed by Dunn’s multiple-comparison test. (C) yfp mRNA abundance for strains bearing the indicated constructs, determined by qPCR and normalized to expression of endogenous sigA. Error bars denote standard deviation. Strains were compared by ANOVA with Tukey’s HSD. (D) Transcripts containing the pmyc1tetO-associated 5′ UTR are translated more efficiently than leaderless transcripts or those containing the sigA 5′ UTR. (E) Published M. tuberculosis mRNA abundance (42) and protein abundance (62) levels for genes that have a single TSS and are leaderless or have 5′ UTRs of ≥15 nt. Protein and mRNA abundance were significantly correlated for both gene structures (P < 0.0001, Spearman’s ρ). Linear regression analysis revealed that the slopes were statistically indistinguishable (P = 0.44). (F) The half-lives of yfp mRNA produced from the indicated constructs were measured. Error bars denote 95% CI. Half-lives were compared using linear regression analysis (n = 3). (G) Protein abundance, mRNA abundance, mRNA half-life, and calculated transcript production rate for leaderless transcripts compared to transcripts with 5′ UTRs. Note that some data shown in Fig. 2 and 3 are reproduced here to facilitate comparisons. ****, P < 0.0001; ***, P < 0.001; ns, P > 0.05.