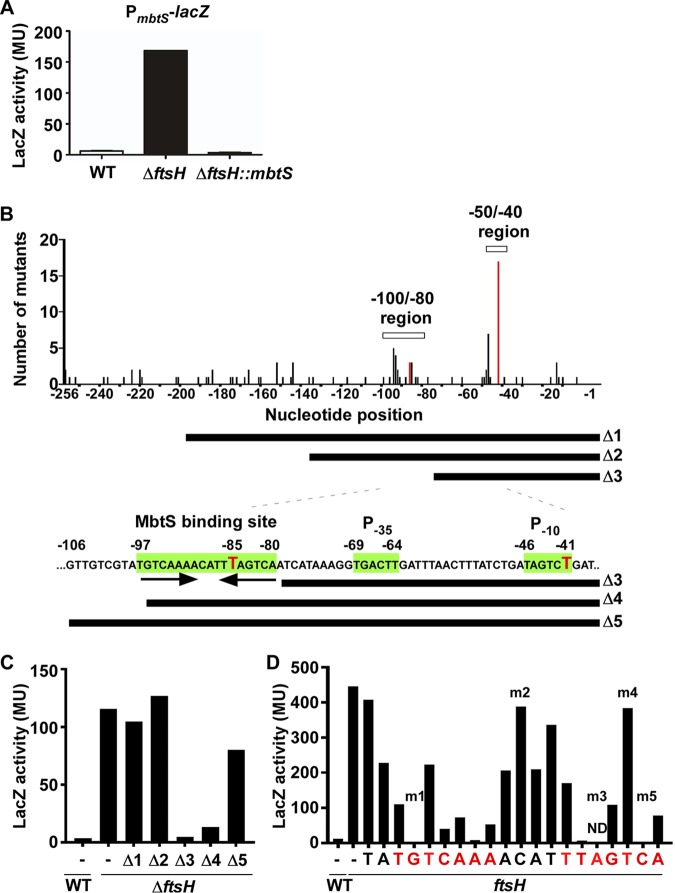
FIG 4.
MbtS recognizes an inverted repeat in its own promoter. (A) Requirement of MbtS for the promoter activity of PmbtS. The PmbtS-lacZ fusion plasmid pOS1-PmbtS-lacZ was inserted into the test strains. Overnight cultures of the test strains were used for the LacZ assay. WT, wild-type strain USA300; ΔftsH, ftsH deletion mutant; ΔftsH::mbtS, ΔftsH with a transposon insertion in mbtS. MU, Miller units. The experiment was carried out in triplicate and repeated twice with similar results. (B) Mutational analyses of PmbtS. The PmbtS sequence in pOS1-PmbtS-lacZ was randomly mutated by error-prone PCR. The 105 mutations identified in 43 white colonies were plotted in PmbtS. On the x axis, −1 indicates the upstream nucleotide right next to the ATG start codon. The y axis shows the number of mutations identified. Two regions, −50/−40 and −100/−80, contained a noticeably higher number of mutations than other regions. Two red bars indicate the locations of two single mutations that abolished the PmbtS activity. The bars under the graph show the PmbtS sequence remaining in the deletion mutant plasmids (Δ1 to Δ5). The putative promoter elements (P−35 and P−10) and the 18-bp sequence containing an inverted repeat are indicated in green. The red Ts are the nucleotides corresponding to the red bar in the graph above. (C) LacZ activities of the five deletion mutants of PmbtS. WT, wild-type USA300; ΔftsH, ftsH deletion mutant of USA300; −, no mutation. (D) LacZ activities of the strains carrying the Δ5 plasmid with a point mutation in the putative MbtS binding sequence. WT, wild-type strain RN4220; ftsH, RN4220 carrying a transposon insertion in ftsH (RN4220::ftsH). The nucleotides in the inverted repeat are shown in red. The mutants indicated by m1 to m5 were used for further experiments (Fig. 5). ND, not determined.