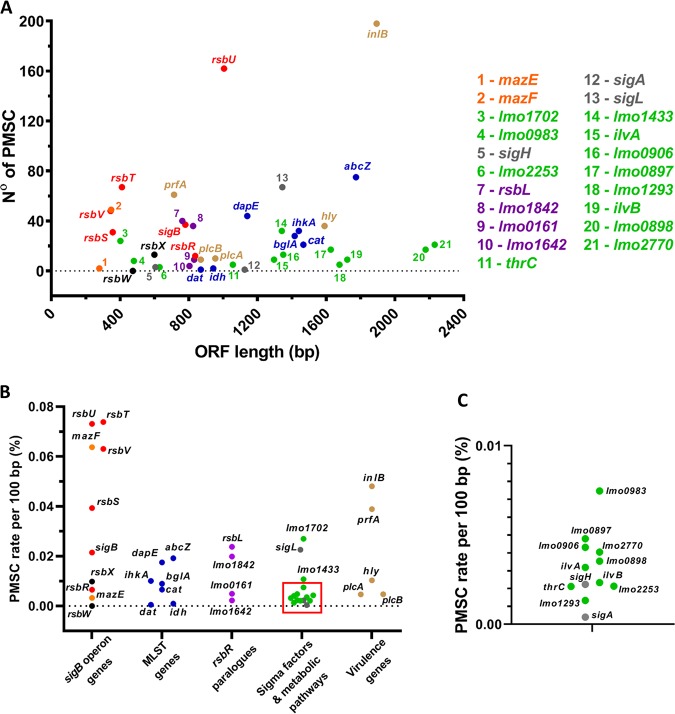
FIG 8.
Occurrence of PMSCs is elevated in genes encoding positive effectors of σB activity. Shown are results for PMSC mutations retrieved from the in silico analysis of the 22,340 genomes of L. monocytogenes strains available. (A and B) The total number of identified PMSCs by ORF length (A) and the PMSC rate normalized by 100 bp of the ORF length (B) for genes comprising the sigB operon and characterized as putative positive regulators of σB activity (red), the putative σB negative regulators rsbX and rsbW (black), and the mazEF genes (orange). MLST genes are in blue, and rsbR paralogues are in purple. The housekeeping gene sigA and the sigma factor genes sigL and sigH are in gray. Two genes downstream of the sigB operon, lmo0897 and lmo0898, and genes encoding metabolic pathways of glutathione (lmo1702, lmo0983, lmo1433, lmo0906, and lmo2770), glycerol (lmo1293), threonine (thrC), isoleucine (ilvA and ilvB), and glucose (lmo2253) are in green, and prfA, inlB, hly, plcA, and plcB are in brown. (C) Expanded area indicated by the square in panel B.