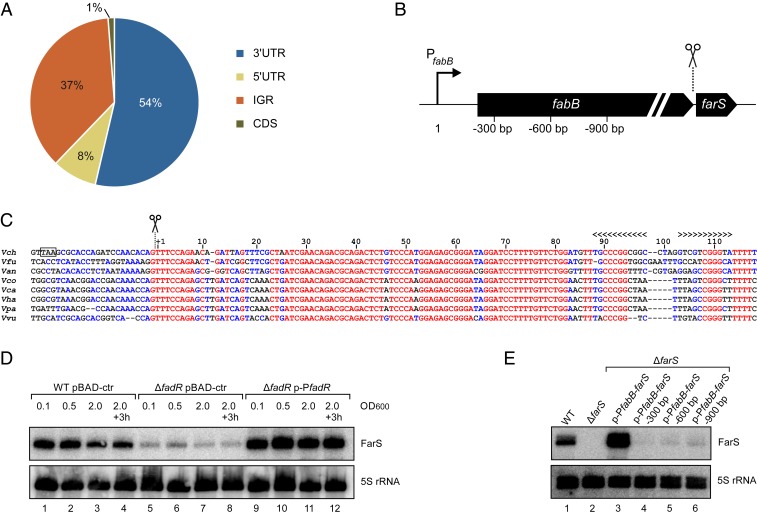
Fig. 2.
Identification and expression of the FarS sRNA. (A) Classification of Hfq-binding sRNAs according to their genomic location. The pie chart shows the relative fractions of Hfq-binding sRNAs (fold enrichment > 2, P value ≤ 0.05) originating from 3′UTRs, intergenic regions (IGRs), 5′UTRs, and coding sequences (CDSs). (B) Schematic representation of the fabB-farS genomic organization. Scissors indicate the processing site. Numbers correspond to the fabB promoter truncations tested in E. (C) Alignment of farS sequences in different Vibrio species. The sequences were aligned using the Multalign tool (76). The start of the sRNA and the Rho-independent terminator are indicated. The stop codon of fabB in V. cholerae is marked with a black box. Vch, Vibrio cholerae; Vfu, Vibrio furnissii; Van, Vibrio anguillarum; Vco, Vibrio coralliilyticus; Vca, Vibrio campbellii; Vha, Vibrio harveyi; Vpa, Vibrio parahaemolyticus; Vvu, Vibrio vulnificus. (D) V. cholerae wild-type and ΔfadR cells harboring either a control plasmid (pBAD-ctr) or a plasmid containing the fadR gene and its native promotor (p-PfadR) were cultivated in LB medium. Total RNA samples were collected at different stages of growth, and expression of FarS was analyzed on Northern blot. 5S rRNA was used as loading control. (E) V. cholerae wild-type and ΔfarS strains harboring different plasmids containing fabB-farS gene fragments (as indicated in B) were grown to stationary phase (OD600 of 2.0) in LB medium. Northern blot analysis was performed to determine FarS levels. Probing for 5S rRNA served as a loading control.