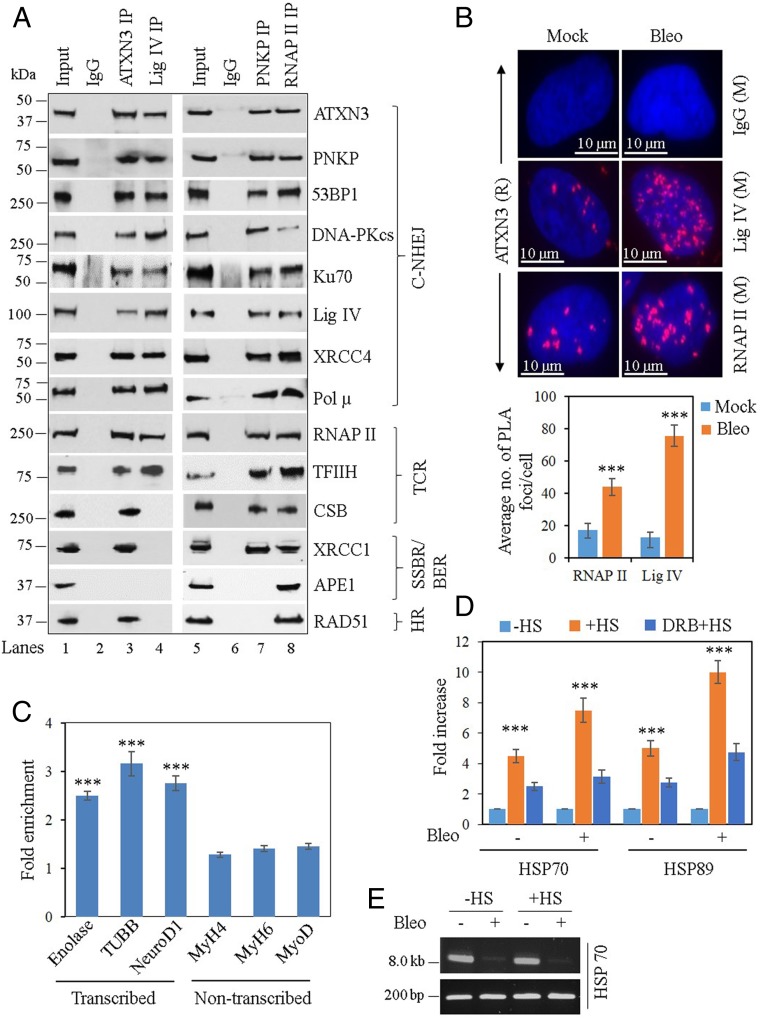
Fig. 1.
Partial characterization of DSB repair complexes and ATXN3’s association with transcribed vs. nontranscribed genes. (A) Benzonase-treated NEs from WT mouse cerebellum were immunoprecipitated with anti-ATXN3 (lane 3), anti-Lig IV (lane 4), anti-PNKP (lane 7), anti-RNAP II (lane 8) Abs, or control IgG (mouse; lane 2 and rabbit; lane 6), and tested for the presence of associated proteins using specific Abs. One representative figure is shown (n = 3). BER, base excision repair; TCR, transcription-coupled repair. (B, Upper) Detection of ATXN3′s (anti-rabbit [R] Ab) association with Lig IV and RNAP II (both anti-mouse [M] Abs) in mock or Bleo-treated NSCs by proximity ligation assay (PLA). Nuclei were counterstained with DAPI (blue) and nonspecific Ab (IgG, mouse) was used as control. Scale bar: 10 µm. (Lower) Average number of proximity ligation assay foci per cell were calculated from 21 randomly selected cells in each sample. Error bars represent ± SD of the mean. The data were significant at ***P < 0.005 between mock and Bleo treatment (n = 3). (C) ChIP was performed from WT mouse cerebellum (n = 3) with anti-ATXN3 Ab and binding to the exonic regions of transcribed (Enolase, TUBB, and NeuroD1) vs. nontranscribed (MyH4, MyH6, and MyoD) genes was quantified (as fold-enrichment = percent input normalized over IgG) by qPCR from immunoprecipitated DNA. Error bars represent ± SD of the mean. ***P < 0.005, showing statistical significance between a particular transcribed gene and all of the nontranscribed genes. (D) SH-SY5Y cells were either mock-treated (−HS) or subjected to heat shock (+HS), or DRB treatment followed by HS (DRB + HS). The cells were further mock- (−) or Bleo- (+) treated and ChIP was performed with anti-ATXN3 Ab or control IgG. Binding to the HSP70 and HSP89 genes was quantified by qPCR from immunoprecipitated DNA. Percent input over IgG was calculated and represented as fold increase with −HS samples arbitrarily considered as unity. Error bars represent ± SD of the mean (n = 3). The data were significant at ***P < 0.005 between each set of −HS/+HS and +HS/DRB+HS. (E) LA-qPCR–mediated estimation of DNA damage in the HSP70 gene before (−HS) and after heat-shock (+HS). A short genomic fragment (∼200 bp) was amplified to normalize the LA-qPCR data.