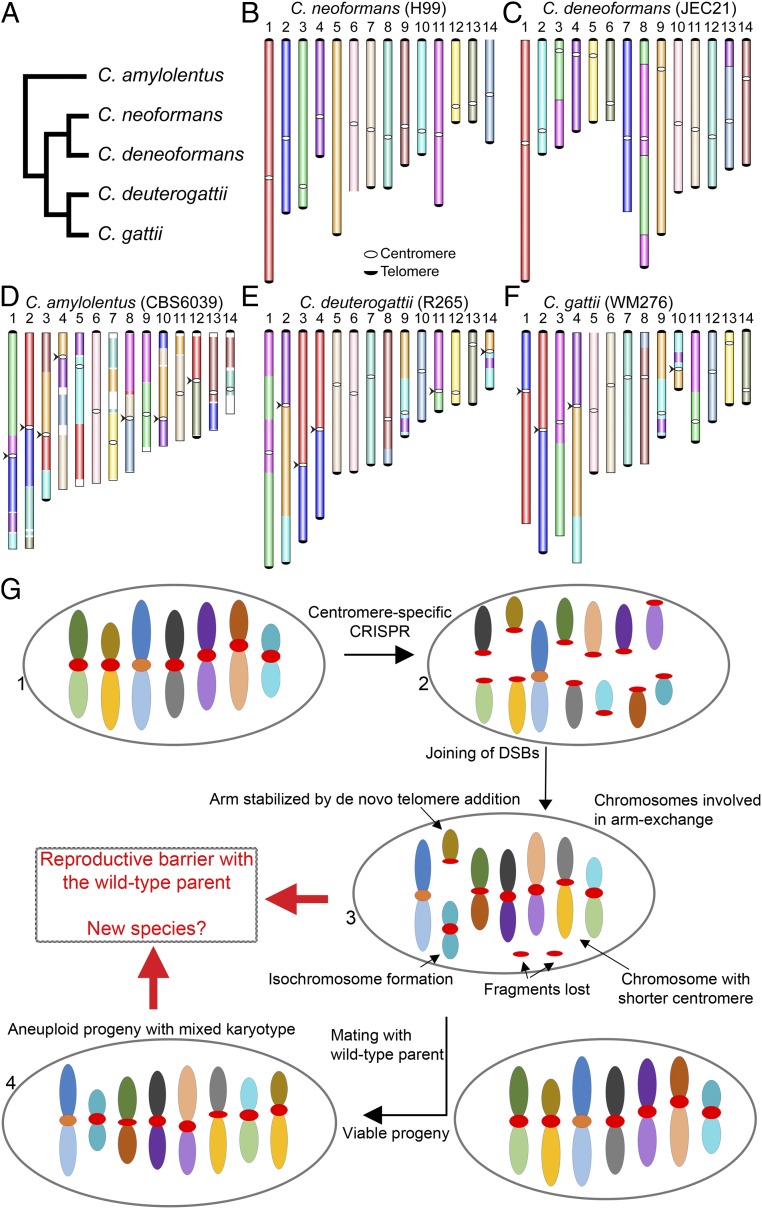
Fig. 7.
A model proposing the evolution of reproductive isolation induced by centromere breaks. (A) A schematic representing phylogeny showing the relationships of five Cryptococcus species, for which chromosome-level genome assemblies are currently available and published. (B–F) Chromosome maps for five species: C. neoformans, C. deneoformans, C. amylolentus, C. deuterogattii, and C. gattii. The synteny maps for all species were generated with C. neoformans as the reference and are colored accordingly. Chromosomal translocations involving centromeres are marked with arrowheads. (G) A model showing the events observed in the study. DSBs generated using CRISPR at centromeres (step 2) reshapes the karyotype following complex repair events. These complex events include the loss of centromere DNA, isochromosome formation, and de novo telomere formation (step 3), similar to what is observed during the process of chromothripsis. The new karyotype can generate a reproductive barrier with the parental isolate and lead to speciation. On the other hand, the strain with the rearranged karyotype could mate with a wild-type isolate, albeit at low frequency, leading to aneuploid progeny (step 4), which can independently establish itself as a new species.