Significance
Prions are self-templating protein conformations that form amyloid fibrils, which are highly ordered protein aggregates. Prions are not only associated with mammalian neurodegenerative diseases but also utilized to switch protein functions, as exemplified by yeast prions. We employed high-speed atomic force microscopy (HS-AFM) to determine how the amyloid fibrils elongate from the monomeric state, since HS-AFM can observe protein dynamics at subsecond and submolecular resolutions. We used the intrinsically disordered region of a yeast prion protein (Sup35NM) as a model, and successfully visualized the conversion of Sup35NM from monomers to oligomers and fibrils. This analysis not only provides direct evidence for the monomer addition to the amyloid fibrils, but also sheds light on the relationship between oligomers and fibrils.
Keywords: yeast prion, amyloid, high-speed AFM, intrinsically disordered proteins, sup35
Abstract
The yeast prion protein Sup35, which contains intrinsically disordered regions, forms amyloid fibrils responsible for a prion phenotype [PSI+]. Using high-speed atomic force microscopy (HS-AFM), we directly visualized the prion determinant domain (Sup35NM) and the formation of its oligomers and fibrils at subsecond and submolecular resolutions. Monomers with freely moving tail-like regions initially appeared in the images, and subsequently oligomers with distinct sizes of ∼1.7 and 3 to 4 nm progressively accumulated. Nevertheless, these oligomers did not form fibrils, even after an incubation for 2 h in the presence of monomers. Fibrils appeared after much longer monomer incubation. The fibril elongation occurred smoothly without discrete steps, suggesting gradual conversions of the incorporated monomers into cross-β structures. The individual oligomers were separated from each other and also from the fibrils by respective, identical lengths on the mica surface, probably due to repulsion caused by the freely moving disordered regions. Based on these HS-AFM observations, we propose that the freely moving tails of the monomers are incorporated into the fibril ends, and then the structural conversions to cross-β structures gradually occur.
Amyloidogenic proteins alter their conformations to form amyloid fibrils composed of cross-β structures. The amyloid fibrils are associated with numerous pathologies, including mammalian neurodegenerative diseases such as Alzheimer’s and infectious prion diseases (1, 2). The growth of amyloid fibrils progresses by a self-templating dock-lock mechanism. First, amyloidogenic protein molecules associate with each other to form a nucleus with a distinct conformation. The nucleus then incorporates monomers or oligomers and converts their protein conformation into an amyloid form (3–5). In addition to amyloids, a variety of oligomers that are often highly toxic to neuronal cells are formed (6–8).
Many amyloidogenic proteins have been identified in the budding yeast Saccharomyces cerevisiae (9). They are responsible for the propagation of non-Mendelian genetic traits, called yeast prions (10–12). Among them, the best studied yeast prion protein is Sup35, which forms amyloids causative of [PSI+], one of the prion phenotypes (13). The monomeric form of Sup35 functions as a translation termination factor (eRF3) (10–12). The function of Sup35 as eRF3 is impaired by the conversion of the Sup35 conformation to amyloids in the [PSI+] cells, resulting in translation read-through at the stop codons (10–12). The [PSI+] phenotypes are inherited in a non-Mendelian manner because the amyloids are transmitted to progeny cells, in which the newly synthesized Sup35 is incorporated into the amyloids in a self-propagating manner (11, 12, 14).
Amyloid formation by Sup35 depends on the glutamine/asparagine (Q/N)-rich N-terminal (N) domain containing 5.5 imperfect repeats (PQGGYQQYN) (15–17), whereas the highly charged middle (M) domain has strong solubilizing activity (10, 11, 18–20). The N and M domains (Sup35NM), which are both intrinsically disordered (ID), govern the ability of the Sup35 protein to exist in two conformers, the prion (amyloid) and nonprion (soluble) states. Sup35NM can adopt a variety of distinct amyloid core structures (21), depending on the temperature and surrounding electrolyte species (19, 21–26). There are two conflicting models of the β-strand arrangement. One is the β-helix model, in which parallel β strands separated by ∼0.48 nm are aligned in a helical pattern running around the fibril axis (22, 27, 28). The other is the in-register parallel β-sheet model, in which identical residues are lined up along the long fibril axis (28, 29).
Numerous studies have elucidated the structural and biochemical properties of Sup35NM fibrils. However, the detailed mechanism underlying the conversion of Sup35NM monomers to amyloid fibrils remains elusive. Previous studies have postulated the following features of the oligomerization and fibrillation of Sup35NM: 1) the Sup35NM monomers form at least two oligomeric intermediates with distinct sizes and reactivities to conformation-specific antibodies (8); 2) the temperature-dependent, transient nonnative interactions in the initial nucleus determine the amyloid conformations and resulting prion strains (24); 3) the fibril grows by monomer addition rather than oligomer addition (5); and 4) the fibril growth rate is different between the two ends of the fibril (17, 29). These previous studies were performed using biochemical methods (22), including fluorescence anisotropy (8), small-angle X-ray scattering (24), analytical ultracentrifugation (5), and fluorescence microscopy (17). However, these methods do not allow the simultaneous and high-resolution assessment of the structures and dynamics involved in the oligomerization and fibrillation of Sup35NM.
To overcome this limitation and obtain insight into these dynamic processes, we used high-speed atomic force microscopy (HS-AFM), which allows direct visualization of protein molecules in dynamic action at high spatiotemporal resolution (30–32). HS-AFM observations of Sup35NM unveiled features of Sup35NM dynamics, providing mechanistic insights into amyloid fibril formation.
Results
Unstructured Regions in Monomeric Sup35NM Visualized by HS-AFM.
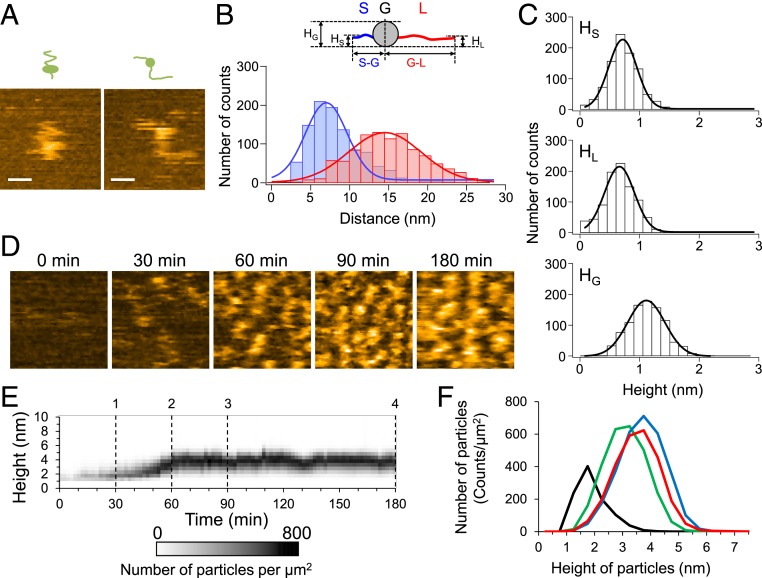
To reveal the structural dynamics of monomeric Sup35NM, we first observed Sup35NM molecules immediately after diluting the urea-denatured protein to a final concentration of 5 nM. The sample was deposited on mica and observed using HS-AFM at 0.05-s intervals at room temperature. Although the movement of the whole molecule was fast and it was difficult to follow the shape of the molecule, the AFM images at high temporal resolution revealed that the Sup35NM molecules contained a globular structure with two highly flexible tail-like structures (Fig. 1A). The rapid fluctuations in the shape of the tail-like structures suggested that both tails are unstructured, consistent with previous reports that Sup35NM contains ID regions (15, 16, 33). The tails had different lengths: the end-to-end distances of the shorter and longer tails were 7.0 ± 3.8 nm and 14.5 ± 6.6 nm, respectively (Fig. 1B). The mean height of the globular part was 1.04 ± 0.44 nm, while those of the short and long tails were 0.64 ± 0.33 nm and 0.59 ± 0.35 nm, respectively (Fig. 1C), which were nearly identical to the value of ∼0.5 nm as measured with HS-AFM for fully disordered proteins (34), confirming again that the Sup35NM tails are unstructured.
Fig. 1.
Structure of Sup35NM monomer and dynamics of oligomer formation. (A) Two representative HS-AFM images of monomeric Sup35NM molecules deposited on mica immediately after denaturant dilution. (Bars, 15 nm; Z-scale, 2.5 nm; imaging rate, 20 fps.) (B) Distributions of end-to-end distance of the shorter (S-G; blue) and longer (G-L; red) tails extending from the globular part. The corresponding blue and red lines indicate most probable Gaussian fitting obtained with 7.0 ± 3.8 nm and 14.5 ± 6.6 nm for mean distance ± SD. (Inset) Schematic of Sup35NM monomer containing two flexible tails and one globular part. HG, HS, and HL represent the heights of the globular part, shorter tail, and longer tail, respectively. (C) Height distributions of the shorter (HS) and longer (HL) tails and the globular part (HG). The corresponding solid lines indicate most probable Gaussian fitting obtained with 0.64 ± 0.33 nm, 0.59 ± 0.35 nm, and 1.04 ± 0.44 nm for mean height ± SD (D) Representative HS-AFM images showing Sup35NM oligomer formation at the indicated times after reaction initiation. (Z-scale, 5 nm; image size, 100 × 100 nm; imaging rate, 5 fps.) (E) Time course of height distribution of Sup35NM particles. (F) Height histograms of Sup35NM monomers/oligomers at 30- (black), 60- (green), 90- (blue), and 180-min (red) incubation times corresponding to dashed lines 1, 2, 3, and 4 in E.
Formation of Sup35NM Oligomers Visualized by HS-AFM.
Immediately after the incubation was started, the Sup35NM molecules (0.5 µM) were invisible when the observation was carried out at a >0.2 s time interval because most of the Sup35NM molecules existed as monomers and moved quickly. However, particles appeared after the Sup35NM was incubated for ∼30 min at room temperature (Fig. 1D). The height of the particles was ∼1.7 nm (Fig. 1 E and F), which was higher than that of the monomers loosely immobilized on the mica surface (Fig. 1C), suggesting that the particles were oligomers. The height of the oligomers gradually increased over the incubation time and reached a plateau at 3 to 4 nm (Fig. 1 D–F). Note that dilution of the solution containing the 3- to 4-nm oligomers from 0.5 to 10 nM did not change the size of the oligomers even after 3-h incubation from the dilution (SI Appendix, Fig. S1A), suggesting that the oligomer formation was irreversible under the condition. In addition, the formation of the 3∼4-nm oligomers was not observed when we performed the observation with a lower concentration (10 nM) of Sup35NM (SI Appendix, Fig. S1 B and C), whereas the Sup35NM monomers at a higher concentration (1.5 µM) rapidly assembled in the oligomers, although the height of the oligomers did not exceed more than 3 to 4 nm even after 3 h (SI Appendix, Fig. S1D). These results suggest that 3∼4-nm oligomer formation was an inherent property of Sup35NM under the condition examined here.
Fibrils did not appear even when 0.5- or 1.5-µM Sup35NM was incubated for 3 h in the AFM sample chamber. The fibril formation took a longer time, typically two to three days, in the AFM sample chamber as well as in solution. Even when we changed the electrostatic property of the mica surface by coating an aminosilane reagent (3-aminopropyltriethoxysilane-mica), we did not find any fibrils after 3 h (SI Appendix, Fig. S2). The property of the substance surface may not affect fibril formation significantly. These results suggest that oligomers do not directly act as seeds for the initiation of fibril formation.
Real-Time Visualization of Sup35NM Fibril Elongation.
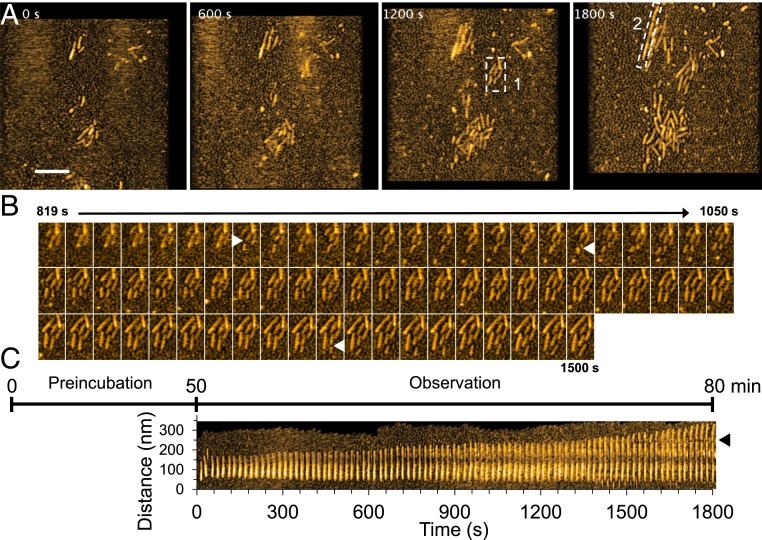
To visualize fibril elongation using HS-AFM, spontaneously formed Sup35NM fibrils, prepared by incubating the Sup35NM solution for more than two days, were used as seeds. When a freshly prepared Sup35NM monomer solution (0.5 µM) was deposited on the preformed Sup35NM fibrils immobilized on the mica surface, fibril elongation was observed immediately within 3 min after monomer deposition (Movie S1). Since durations less than ∼3 min are not long enough for oligomer formation from monomers, as described previously (Fig. 1), this immediate elongation strongly suggested that the fibrils grew by monomer addition, as previously reported (5). The growing fibrils were fragile and occasionally fragmented during HS-AFM imaging, providing new seeds for fibril elongation (Fig. 2 A–C). A kymograph of fibril elongation revealed the strong growth polarity: the elongation mainly occurred at one end of the fibrils (Fig. 2C), consistent with previous studies (17, 29). The growth rate of the fibril shown in Fig. 2C was not constant, and the fibril elongated at a faster rate after ∼70 min. However, a statistical analysis revealed that there is no significant difference between the overall growth rates of fibrils at 0 to 30, 30 to 60, and 60 to 90 min after the addition of fresh monomers (SI Appendix, Fig. S3).
Fig. 2.
HS-AFM images showing fibril growth. (A) Representative HS-AFM images of growing Sup35NM fibrils captured after 50-min preincubation on mica. (Z-scale, 10 nm; bar, 200 nm; imaging rate, 0.1 fps.) (B) Successive HS-AFM images showing seed-dependent fibril growth and fragmentation. The images are those clipped for the region highlighted by dashed box 1 in A. The fragmented positions are indicated by triangles. (C) Kymograph of a representative growing Sup35NM fibril in the region highlighted by dashed box 2 in A. The fragmented positions are indicated by a triangle. The time course in the figure shows the elapsed time including preincubation time since the monomers were added on the fibrils or seeds fixed on mica.
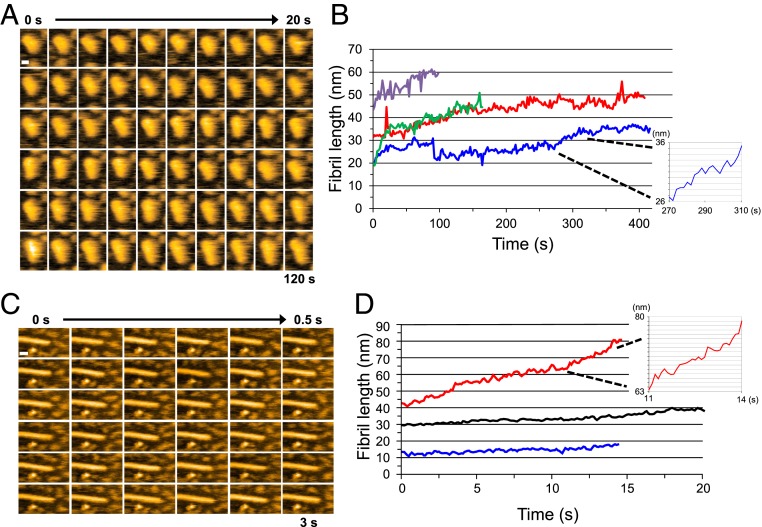
To elucidate the mechanism of fibril growth in more detail, we observed the growth at a higher spatiotemporal resolution (Fig. 3). As shown in Fig. 3 A and B, the fibril elongated smoothly without discrete steps, even at this high spatial resolution (pixel size 0.5 nm), suggesting that after a monomer is incorporated into a fibril end, it is gradually converted into a part of the fibril structure. We also observed the smooth elongation of fibrils even when we added Sup35NM samples containing oligomers; that is, solutions of Sup35NM (0.5 µM) preincubated for 30 and 90 min were added to the preformed fibrils (Fig. 3B). Note that the ∼1.7-nm and the 3∼4-nm oligomers were formed during these preincubation periods, respectively. The smooth elongation again confirmed that fibril elongation occurs by monomer addition rather than oligomer addition. Even when the imaging rate was increased to 10 frames per second (fps), we did not observe any stepwise elongation of the fibrils (Fig. 3 C and D), again confirming that fibril elongation does not occur by oligomer addition.
Fig. 3.
Fibril elongation observed at high spatial and temporal resolution. (A) Successive HS-AFM images of a growing Sup35NM fibril captured at a pixel size of 0.5 nm. (Imaging rate, 0.5 fps; bar, 10 nm.) (B) Time courses of length change of growing Sup35NM fibrils. The different lines indicate different fibrils. Red and blue lines show the time courses of fibril elongation when a Sup35NM sample (0.5 µM) was preincubated for 30 min; green and purple lines show cases when the same sample was preincubated for 90 min. (Inset) Magnified time course taken from the blue line in the period from 270 to 310 s. (Grid interval of y-axis, 1 nm.) (C) Successive HS-AFM images of a growing Sup35NM fibril captured at high temporal resolution. (Imaging rate, 10 fps; bar, 20 nm; pixel size, 1 nm.) (D) Time course of length change of growing Sup35NM fibrils. The different lines correspond to different fibrils. (Inset) Magnified time course taken from the red line in the period from 11 to 14 s. Sup35NM samples (0.5 µM) were preincubated for 30 min. (Grid interval of y-axis, 1 nm.)
Gap between Sup35NM Oligomers and Fibrils.
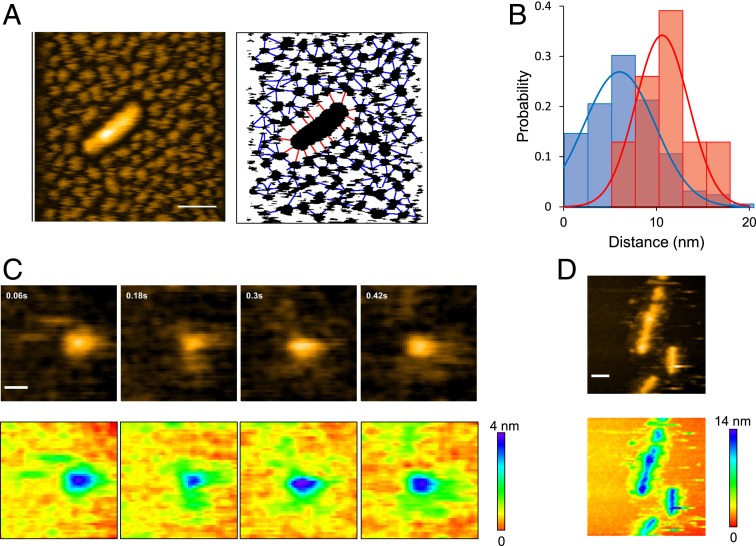
During our HS-AFM observations, we noticed that the oligomers were interspaced with a similar size of gaps, suggesting that they are mutually repulsive (Fig. 4A). Interspace gaps were also observed between a fibril and the oligomers surrounding it. The gap size between adjacent oligomers was distributed around ∼6 nm, while that between a fibril and its neighboring oligomers was distributed around ∼10 nm (Fig. 4B). Since monomeric Sup35NM has disordered regions (Fig. 1), the observed gaps were likely to have originated from the flexible regions of Sup35NM extending out from the oligomers and fibrils. This distinct difference in gap size distribution suggests that the disordered regions extending from the fibril core structures are longer than those from the oligomers. Note that the ionic strength of the solution used in these experiments was relatively high (∼175 mM). The Debye Hückel distance at this ionic strength is ∼0.72 nm, much shorter than the gap distances. Therefore, it is very unlikely that electrostatic repulsion between the particles is the cause of this interparticle spacing. We also confirmed that the interparticle spacing did not depend on the salt concentration (SI Appendix, Fig. S4).
Fig. 4.
Gap between interoligomers and fibril-oligomers. (A, Left), HS-AFM image showing a Sup35NM fibril densely surrounded by Sup35NM oligomers. (Z-scale, 15 nm; bar, 50 nm; imaging rate, 0.2 fps.) (A, Right), Binarized image corresponding to the image on the Left, together with lines connecting neighboring oligomers (blue) and those connecting the fibril and surrounding oligomers (red). (B) Distributions (bars) and Gaussian fits (lines) of the gap distances between neighboring oligomers (blue) and between the fibril and its surrounding oligomers (red) shown in A, Right. The Gaussian fits were obtained with 6.0 ± 3.8 nm and 10.6 ± 2.9 nm for the interoligomer and the fibril-oligomer distances, respectively. The single-Gaussian distributions are statistically significant (P < 5 × 10−6 by t test analysis). (C, Upper), HS-AFM images of a Sup35NM oligomer; Sup35NM concentration, 10 nM. (Image size, 50 nm × 50 nm; imaging rate, 16.7 fps; Z-scale, 4 nm; bar, 10 nm.) (C, Lower), Height displays in rainbow color for the images shown in C, Upper. (D, Upper) HS-AFM image of Sup35NM fibrils with some structures extending from the lateral side. (Image size, 200 nm × 200 nm; imaging rate, 1 fps; Z-scale, 14 nm; bar, 30 nm.) (D, Lower), Height display in rainbow color for the HS-AFM image shown in D, Upper.
To determine whether these gaps originate from the disordered regions extending from the oligomers and the fibril core, we next observed a single oligomer at higher temporal resolution at 0.06-s intervals. As in the case of monomers, we observed freely moving unstructured regions extending from the oligomer (Fig. 4C and Movie S2), suggesting that the interparticle gaps observed with low temporal resolution imaging were likely to be occupied by unstructured regions that would not be visible with such slow imaging. We also observed some structures outside the lateral sides of the fibrils (Fig. 4D). These structures were visible even with 1-s–interval imaging, indicating that the structures extending out of the fibrils were relatively solid and not freely moving. Notably, such structures were not observed at the fibril ends. We also confirmed by 0.06-s–interval imaging that the moving oligomers were very close to the fibril end, almost interacting with the fibril end in some snapshots, although incorporation of oligomers into the fibril end was not observed (SI Appendix, Fig. S5A and Movie S3). Unlike the fibril ends, no oligomers approached the lateral side of the fibrils, indicating that some structure denies oligomer access to this side of the fibrils (SI Appendix, Fig. S5B and Movie S4). It might play an important role in the straight growth of fibrils without branching.
Discussion
The conversion of monomers to oligomers and the fibril elongation of Sup35NM were observed in this study, using HS-AFM. HS-AFM has been used to visualize the motion of disordered regions in ID proteins (34). In fact, the HS-AFM imaging of monomeric Sup35NM revealed shorter and longer flexible tails extending from the globular core structure. Predictor of natural disordered regions (PONDR) (35) revealed that Sup35NM has disordered regions, including the N-terminal ∼80 and C-terminal ∼125 amino acids (SI Appendix, Fig. S6), indicating that the shorter tail would be the N-terminal ∼80 residues in Sup35NM, followed by the core structure. A previous hydrogen/deuterium exchange experiment with Sup35NM fibrils showed that residues 76 through 100 in Sup35NM are largely unprotected in the fibrils (23). Therefore, the core structure in the monomeric Sup35NM visualized by HS-AMF might not be incorporated into the fibrils.
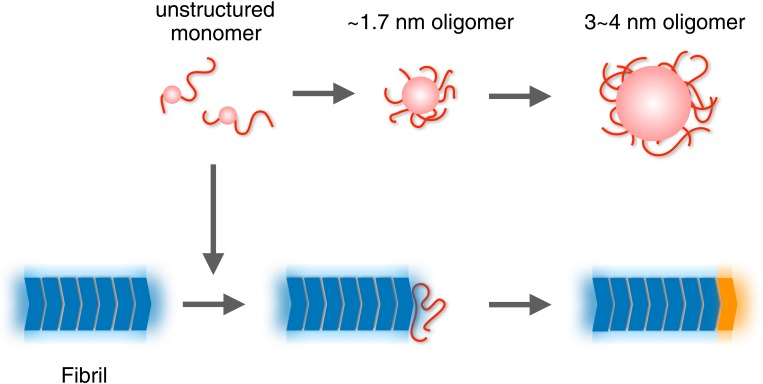
Previous studies demonstrated that Sup35NM forms nonamyloid oligomers (5, 24, 36). Our HS-AFM observations are consistent with these studies, having unveiled the detailed properties of the oligomers. In particular, HS-AFM allowed direct observation of the kinetics of oligomer formation from monomers. Based on the HS-AFM data, we propose the following mechanism of fibril formation (Fig. 5). Starting from monomers, Sup35NM forms the 3∼4-nm oligomers via the intermediate ∼1.7-nm oligomers. Importantly, neither oligomer is a direct precursor of fibril formation, for the following reasons. First, without any seed from preformed fibrils, fibril formation and elongation do not occur in the mixture containing those oligomers for at least ∼3 h under the conditions tested here. Second, preformed fibril-nucleated elongation instantaneously occurs even when the oligomers have not yet accumulated, implying that monomers, not oligomers, are the direct precursor of fibril elongation, as previously reported (5). Our HS-AFM observations also suggest that the final conversion to the core structure of a fibril occurs gradually. What is the physiological role of the oligomers formed from Sup35NM? ID regions are known to mediate the assembly of both hetero-complexes and homo-oligomers, which are often involved in cellular signaling and regulation (37). Therefore, the ID-mediated oligomers of Sup35NM might participate in a cellular process different from the original function of Sup35 as the translation termination factor. Recently, Sup35NM was found to have a physiological function, besides amyloid formation, which is forming a droplet via pH-regulated phase separation under stress (38). At present, the relationship between the oligomers we observed and the droplet in the stressed cells is unclear, but it should be investigated in the future.
Fig. 5.
Schematic model for the formation of Sup35NM oligomers and fibrils. Unstructured Sup35NM monomers (red) are converted to ∼1.7-nm oligomers and then to 3∼4-nm oligomers. Fluctuating disordered structures are depicted as tail-like shapes. Globular and oligomer core parts are shown as spherical shapes. Core structures of fibrils (dark blue) are surrounded by some mobile structures (pale blue). The unstructured monomer is incorporated into a growing end of the fibrils. The attached unstructured monomer is conformationally converted to a core to become a newly elongated part of the fibril (orange).
The HS-AFM observations in this study provided several insights into the interspatial gaps among oligomers and fibrils. As shown in Fig. 4, the Sup35NM oligomers remained dispersed from each other and from the fibrils. The distribution of the interoligomer separations is distinctly different from the separations between the fibrils and oligomers. The extended disordered structures in the oligomers repulse each other, resulting in gap space formation. The gap induced by the disordered regions is considered to be the reason that oligomers are highly unlikely to become a nucleus, although the structure of the nuclei as seeds still remains unclear. Interestingly, the structures outside the fibrils were relatively solid and not observed at the fibril ends. This could be the mechanism underlying the straight elongation of fibrils without branching and thickening.
Materials and Methods
Preparation of Sup35NM.
The purification of Sup35NM, with a histidine tag and a cysteine at the C terminus (17), was performed as described previously, with several modifications (17). Sup35NM was produced in Escherichia. coli BL21(DE3), with expression induced for 2 h with 1-mM isopropyl β-D-1-thiogalactopyranoside at 37 °C. Sup35NM was suspended in 20-mM sodium phosphate (pH 7.4), 500-mM NaCl, 8-M urea, 10-mM imidazole, and 10-mM 2-mercaptoethanol. The Sup35NM solution was applied to a Ni-NTA Superflow (Qiagen) column, eluted with 500-mM imidazole. The fraction containing Sup35NM was dialyzed against 0.1% (vol/vol) trifluoroacetic acid and subjected to reversed-phase high-pressure liquid chromatography (Poros R2/10). The column was equilibrated with 0.1% (vol/vol) trifluoroacetic acid and washed with 15% acetonitrile in 0.1% trifluoroacetic acid. Sup35NM was eluted with a 15 to 40% (vol/vol) acetonitrile gradient in 0.1% trifluoroacetic acid, and fractions containing Sup35NM were collected.
HS-AFM Observation.
The HS-AFM setup, equipped with small cantilevers (BL-AC10-DS, Olympus: spring constant, ∼0.1 N/m; resonance frequency, 400 to 500 kHz in water), was operated in tapping mode (39, 40). An amorphous carbon tip was grown on the top of the original tip by electron-beam deposition, using a field emission scanning electron microscope (ERA8000-FE, Elionix). The cantilever free oscillation amplitude was set at ∼1.5 nm, and the set point amplitude was adjusted at 80 to 90% of the free oscillation amplitude so that the oscillation energy would be lost by 2 to 4 kBT by tip-sample contact (kB, Boltzmann constant; T, room temperature in kelvin). To observe Sup35NM monomers containing ID regions, 8-M urea-denatured Sup35NM (50 μM) was diluted to 5 nM with 5-mM sodium phosphate (pH 7.0). For the observation of oligomer formation, 8-M urea-denatured Sup35NM (50 μM) was diluted to 0.5 μM with Buffer A containing 5-mM potassium phosphate (pH 7.3), 150-mM NaCl, 25-mM KCl, and 1-mM DTT, followed by incubation at room temperature. For the observation of fibril growth, preformed Sup35NM fibrils prepared by incubating Sup35NM monomers for 2 d at room temperature (25 °C) were deposited on a mica stage. After this sample stage was introduced into the HS-AFM sample chamber, the chamber was immersed in Buffer A containing 0.5-μM monomeric Sup35NM. The obtained successive HS-AFM images were processed and analyzed by ImageJ software.
Statistical Analysis.
The Gaussian fits of the histograms from length and height measurements were obtained with mean ± SD. A two-tailed t test was performed in the statistical analysis of the gap between the interoligomer and the fibril-oligomer and in the analysis of the effect of ionic strength distances between particles and between fibril and particles. P < 0.05 was considered statistically significant.
Data Availability Statement.
Data in this manuscript have been uploaded to the Mendeley Data public repository (https://data.mendeley.com/datasets/yvdxcxrbzw/2).
Supplementary Material
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (JP18H04512, JP18H01837, and JP19H05389 to T.U.; JP19K06596 to H.K.; and 24657070, 24113705, and 26116002 to H.T.) from Japan Society for the Promotion of Science (JSPS) and Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; by grants from the Daiichi Sankyo Foundation of Life Science and the Mochida Memorial Foundation (to H.T.), the Japan Science and Technology Agency (JST) Basic Research program (CREST) program (JPMJCR13M1 to T.A. and H.K.; JPMJCR1762 to N.K. and H.K.); and the World Premier International Research Center Initiative (WPI), MEXT, Japan, and the Kanazawa University CHOZEN project.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Data in this manuscript have been uploaded to the Mendeley Data public repository (https://data.mendeley.com/datasets/yvdxcxrbzw/2).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916452117/-/DCSupplemental.
References
- 1.Knowles T. P., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. B., Biology and genetics of prions causing neurodegeneration. Annu. Rev. Genet. 47, 601–623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon M. J., Williams A. D., Wetzel R., Myszka D. G., Kinetic analysis of beta-amyloid fibril elongation. Anal. Biochem. 328, 67–75 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Esler W. P., et al. , Alzheimer’s disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry 39, 6288–6295 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Collins S. R., Douglass A., Vale R. D., Weissman J. S., Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2, e321 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S., et al. , Soluble oligomers of amyloid Beta protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron 62, 788–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haass C., Selkoe D. J., Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Krishnan R., et al. , Conserved features of intermediates in amyloid assembly determine their benign or toxic states. Proc. Natl. Acad. Sci. U.S.A. 109, 11172–11177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alberti S., Halfmann R., King O., Kapila A., Lindquist S., A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuite M. F., Cox B. S., Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 4, 878–890 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Wickner R. B., Edskes H. K., Shewmaker F., Nakayashiki T., Prions of fungi: Inherited structures and biological roles. Nat. Rev. Microbiol. 5, 611–618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liebman S. W., Chernoff Y. O., Prions in yeast. Genetics 191, 1041–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickner R. B., [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 264, 566–569 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Taguchi H., Kawai-Noma S., Amyloid oligomers: Diffuse oligomer-based transmission of yeast prions. FEBS J. 277, 1359–1368 (2010). [DOI] [PubMed] [Google Scholar]
- 15.King C. Y., et al. , Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. U.S.A. 94, 6618–6622 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glover J. R., et al. , Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Inoue Y., Kishimoto A., Hirao J., Yoshida M., Taguchi H., Strong growth polarity of yeast prion fiber revealed by single fiber imaging. J. Biol. Chem. 276, 35227–35230 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Shorter J., Lindquist S., Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6, 435–450 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S., Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Uptain S. M., Lindquist S., Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56, 703–741 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Tanaka M., Collins S. R., Toyama B. H., Weissman J. S., The physical basis of how prion conformations determine strain phenotypes. Nature 442, 585–589 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Krishnan R., Lindquist S. L., Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyama B. H., Kelly M. J., Gross J. D., Weissman J. S., The structural basis of yeast prion strain variants. Nature 449, 233–237 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Ohhashi Y., Ito K., Toyama B. H., Weissman J. S., Tanaka M., Differences in prion strain conformations result from non-native interactions in a nucleus. Nat. Chem. Biol. 6, 225–230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh V., et al. , The Hofmeister effect on amyloid formation using yeast prion protein. Protein Sci. 19, 47–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubin J., et al. , Ion-specific effects on prion nucleation and strain formation. J. Biol. Chem. 288, 30300–30308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishimoto A., et al. , beta-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem. Biophys. Res. Commun. 315, 739–745 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Frederick K. K., et al. , Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register. Proc. Natl. Acad. Sci. U.S.A. 114, 3642–3647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePace A. H., Weissman J. S., Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat. Struct. Biol. 9, 389–396 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Kodera N., Yamamoto D., Ishikawa R., Ando T., Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 468, 72–76 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Ando T., Uchihashi T., Kodera N., High-speed AFM and applications to biomolecular systems. Annu. Rev. Biophys. 42, 393–414 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Ando T., High-speed AFM imaging. Curr. Opin. Struct. Biol. 28, 63–68 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay S., Krishnan R., Lemke E. A., Lindquist S., Deniz A. A., A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. U.S.A. 104, 2649–2654 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyagi A., et al. , Visualization of intrinsically disordered regions of proteins by high-speed atomic force microscopy. Chemphyschem 9, 1859–1866 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Obradovic Z., et al. , Predicting intrinsic disorder from amino acid sequence. Proteins 53 (suppl. 6), 566–572 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Serio T. R., et al. , Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maharana S., et al. , RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchihashi T., Kodera N., Ando T., Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Ando T., et al. , A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 98, 12468–12472 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data in this manuscript have been uploaded to the Mendeley Data public repository (https://data.mendeley.com/datasets/yvdxcxrbzw/2).