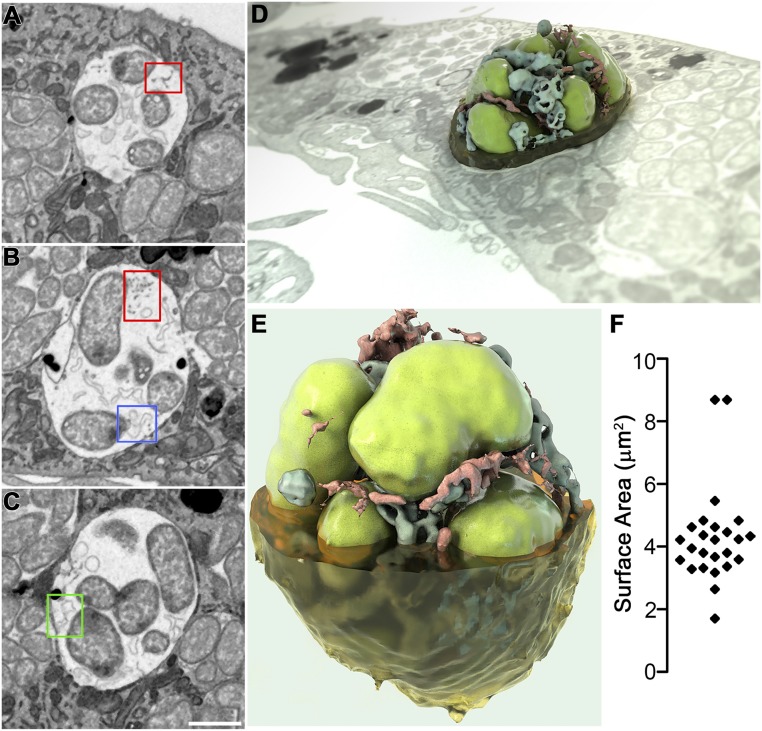
Fig. 7.
Three-dimensional imaging of intraluminal structures in E. chaffeensis-containing inclusions by FIB-SEM. E. chaffeensis-infected DH82 macrophages at 2 dpi were embedded in resin and subjected to iterative milling with a focused ion beam followed by imaging by SEM. For 3D imaging, a region of interest (red box in Movie S2) through one inclusion was selected and segmented. Image data were binned three times in the xy plane to give a final voxel size of 15 × 15 × 15 nm, and individual 2D images were merged, cropped, aligned, and then reconstructed to produce Movie S3. (A–C) Selected 2D slices through one inclusion demonstrated the presence of filaments or vesicles connected to the inclusion membrane (A) or bacterial (B) membrane (red boxes); branching tubules attached at one edge to either the bacterial membrane (B, blue box), or inclusion membrane (C, green box). (Scale bar, 1 μm.) (D and E) Reconstructed 3D images representing a bacterial vacuole inside a cell or viewed at a different angle (3D ultrastructural volume of the sample is shown in Movie S3). Segmentation shows one vacuole with bacteria (green), filaments (red), and vesicles (blue). (F) Scatter plot of the surface area of each individual bacterium in Movie S3 (4 μm2 average).