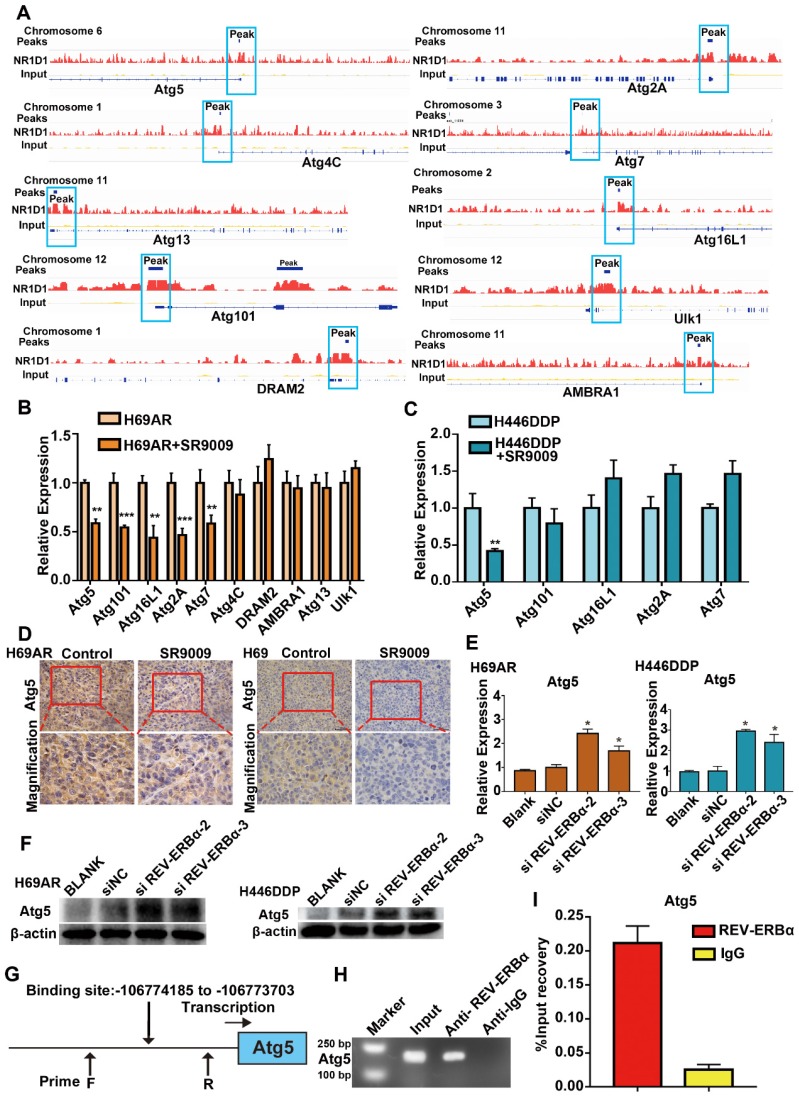
Figure 6.
Core autophagy gene Atg5 was novel and functional REV-ERBα target in SCLC. (A) Analyses of ChIP-sequence data indicated that REV-ERBs peaks were present in core autophagy genes Atg5, Atg101, Atg16L1, Atg2A, Atg7, Atg4C, DRAM2, AMBRA1, Atg13 and Ulk1 (false discovery rate ≤ 0.05). (B) QRT-PCR analysis of autophagy gene Atg5, Atg101, Atg16L1, Atg2A, Atg7, Atg4C, DRAM2, AMBRA1, Atg13 and Ulk1 in H69AR cells. The values of relative control were set to 1. The data are presented as means ± SD of three independent experiments. **P < 0.01; ***P < 0.001. (C) QRT-PCR analysis of autophagy gene Atg5, Atg101, Atg16L1, Atg2A and Atg7 in H446DDP cells. The values of relative control were set to 1. The data are presented as means ± SD of three independent experiments. **P < 0.01. (D) Representative IHC staining of autophagy associate protein Atg5 in subcutaneous xenografts of SR9009-treated group or vehicle group. (E and F) Autophagy core genes Atg5 was upregulated in cells with depletion of REV-ERBα by siRNA. The values of relative control were set to 1. The data are presented as means ± SD of three independent experiments. *P < 0.05. (G) Predicted REV-ERBα binding site and the qPCR primer location in the Atg5 promoter region. (H and I) REV-ERBα ChIP-qPCR assessing REV-ERBα enrichment at the Atg5 promoter. The data are presented as means ± SD of three independent experiments.