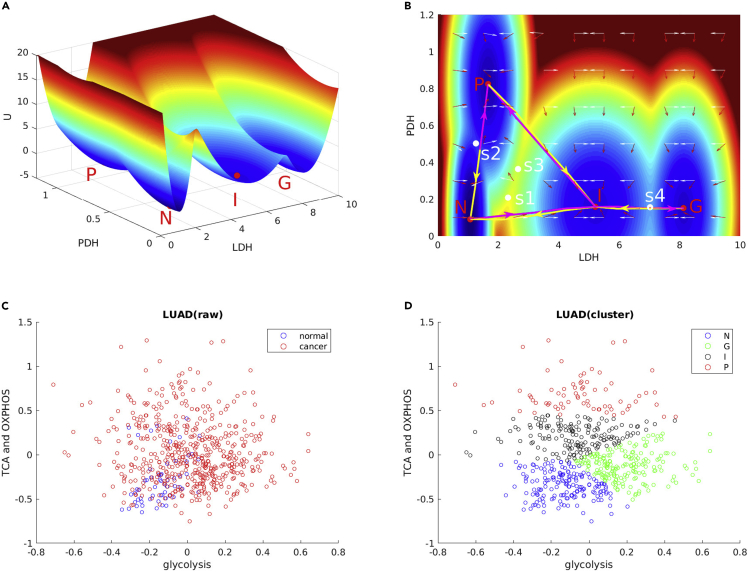
Figure 2.
Landscape of Cancer Gene-Metabolism and Related Gene Expressions from GDC
The landscape is represented in terms of LDH expression level and PDH expression level. N, normal state; P, cancer OXPHOS state; G, cancer glycolysis state; I, cancer intermediate state. s1, saddle between normal state and cancer intermediate state; s2, saddle between normal state and cancer OXPHOS state; s3, saddle between cancer intermediate state and cancer OXPHOS state; s4, saddle between cancer intermediate state and cancer glycolysis state. The yellow arrows represent the paths from N to I, from I to P, from N to P, and from I to G; the magenta arrows represent the paths from I to N, from P to N, from P to I, and from G to I. The white arrows represent the directions of the steady-state probability flux, and the red arrows represent the directions of the negative gradient of the potential landscape.
(A) The landscape of cancer gene-metabolism in 3D.
(B) The landscape of cancer gene-metabolism in 2D.
(C) Gene expression data with normal and cancer samples.
(D) Gene expression data clustered by K-means.