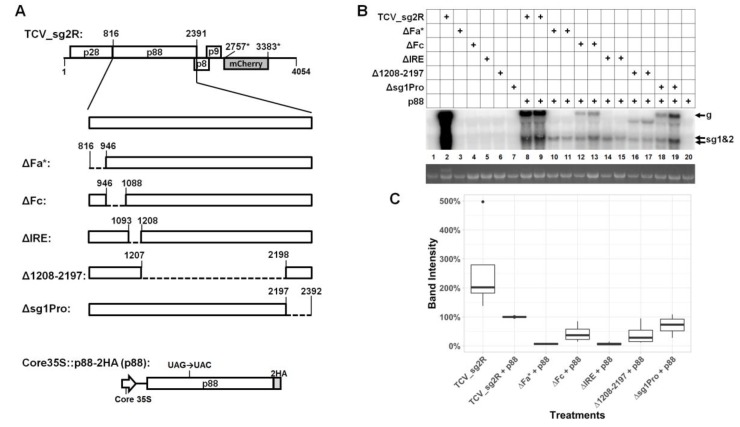
Figure 1.
Deletions within the p88 coding sequence differentially affect the abundance of TCV gRNA in N. benthamiana cells. (A) Schematic representation of five in-frame deletions within the p88 coding sequence. The top diagram depicts the genome of TCV_sg2R, a derivative of TCV encoding mCherry in place of TCV CP. It is the backbone for all deletion and point mutants used in this study. The numbers are nucleotide positions in TCV gRNA. Dashed lines represent the regions deleted. (B) Results of a representative Northern blotting, showing the accumulation levels of different deletion mutants in the presence of trans-supplied p88. None of the mutants could replicate on their own. (C) Quantification of the gRNA accumulation levels of the deletion mutants. The box plots were based on relative intensity of the gRNA bands, derived from four repeats. The band intensity of (TCV_sg2R + p88) was set as the reference for comparison.