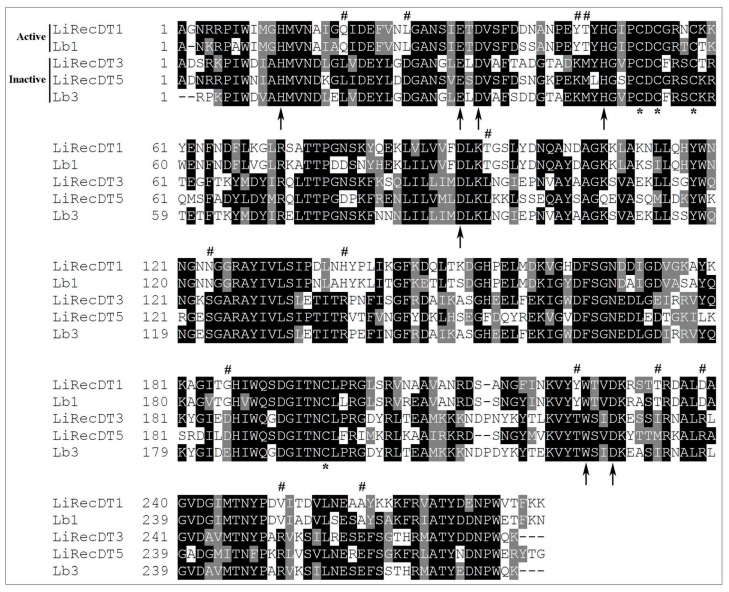
Figure 2.
Enzymatically active and inactive Brown spider venom PLDs. Multiple sequence alignment of phospholipases-D of Loxosceles intermedia (Li) and L. boneti (Lb) venoms: LiRecDT1 (GenBank accession number ABA62021), Lb1 (AY559844)—active PLDs; LiRecDT3 (ABB71184), LiRecDT5 (ABD91847) and Lb3 (AAT66074)—inactive PLDs. Sequences were aligned using the CLUSTAL X2 program [19]. Amino acid identities are shaded in black. Conservative substitutions are in gray, arrows point to amino acid residues involved in catalysis. The asterisks show cysteine residues. Hashtags indicate amino acids in active enzymes that are replaced by amino acids with different side chains in inactive isoforms.