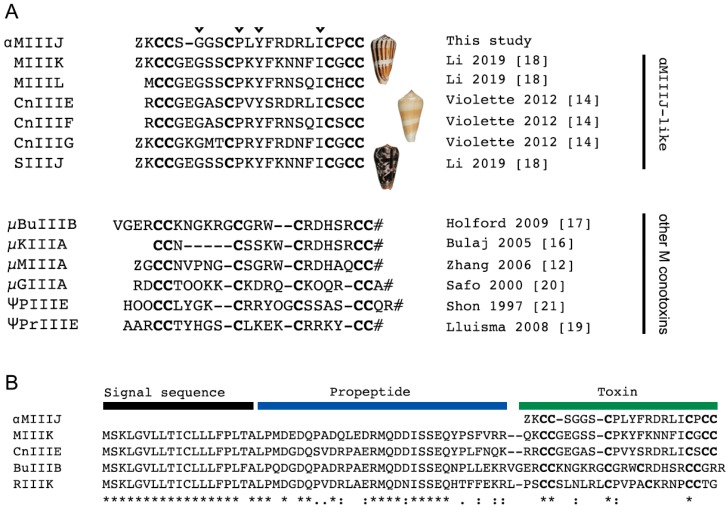
Figure 1.
Sequence of αM-MIIIJ compared with those of other conotoxins. A. Sequence alignment of αM-MIIIJ with related sequences of unknown activity identified in C. magus, C. consors, and C. striatus (top) and M-superfamily conotoxins (bottom) that block V-gated Na channels (μ-prefix) or nAChRs (ψ-prefix for binding at a noncompetitive site). Cysteines are in bold. Identical amino acids are shown with arrowhead on top of αM-MIIIJ sequence. Z: pyroglutamic acid, O: hydroxyproline, #: C-terminal amidation. References [12,14,15,16,17,18,19,20]. B. Precursor sequence alignment of M-superfamily toxins highlights a conserved signal sequence used for toxin gene classification. The precursor sequence of αM-MIIIJ could not be retrieved, but high sequence similarities with αM-MIIIJ-like sequences, including MIIIK (91%), strongly suggest that αM-MIIIJ also belongs to the M-superfamily. Amino acid conservations are denoted by an asterisk (*). Full stops (.) and colons (:) represent a low and high degree of similarity, respectively.