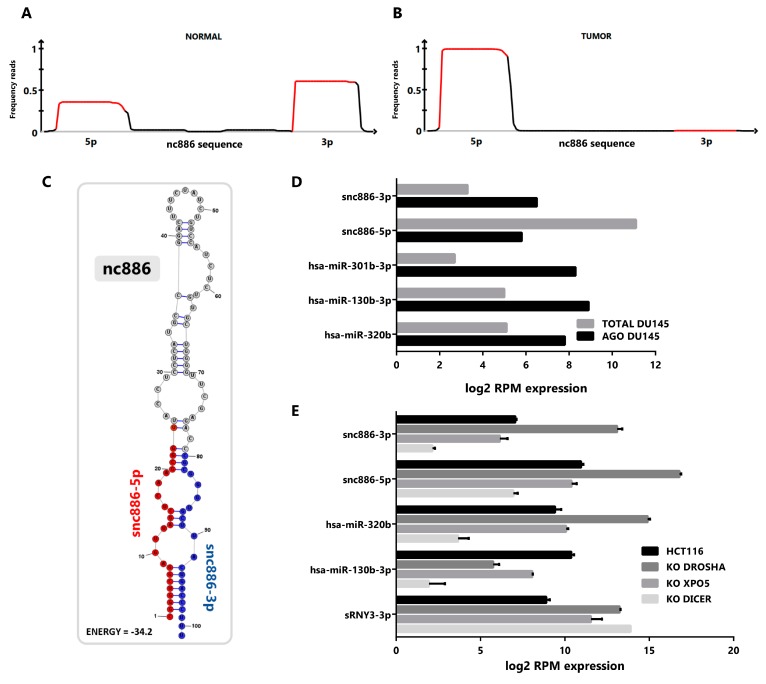
Figure 1.
nc886 derived fragments are produced in prostate cell lines and exhibit microRNA features. (A,B) Mapping of prostate cell small RNA reads along the sequence of nc886. Reads obtained from small-RNA transcriptomes of non-transformed prostate PrEc and PrSC (GEO id: GSE26970) (A) and tumor DU145, PC-3 and LNCaP cell lines (SRA id: SRP109305 and GEO id: GSE66035) (B) were mapped to nc886. A diagram of nc886 (Refseq (NR_030583.2)), plus additional 10 nt at both ends, is depicted below the plots. Red lines in the plots represent the previously annotated hsa-miR-886-3p and -5p. (C) Predicted secondary structure of nc886 (Refseq (NR_030583.2)) based on maximum free energy (MFE) generated with the RNAstructure software [44]. (D) Association of small RNAs to Argonaute in transcriptomic analysis of DU145 cell line. The graph shows the normalized expression, reads per million (RPM), of small non-coding RNAs in total cellular RNA (TOTAL DU145) and in the Argonaute PAR-CLIP fraction (AGO DU145). Data set available at SRA id: SRP075075. (E) Transcriptomic analysis of small non-coding RNAs of the HCT116 cell line with knockouts for the microRNAs biogenesis proteins DROSHA, EXPORTIN 5 and DICER. Data set available at GEO id: GSE77989. Average value and standard error are shown.