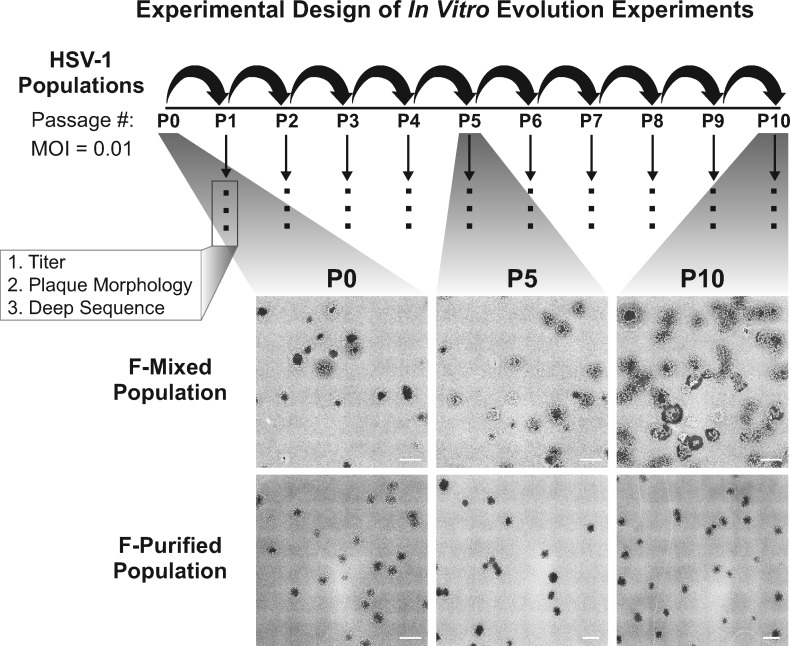
Figure 1.
Schematic of the in vitro evolution experiment approach and observed changes in plaque morphology: two populations of HSV-1 strain F were used to infect Vero cells at a multiplicity of infection (MOI) of 0.01. After infection was allowed to proceed for 72 h, progeny viral populations were harvested. This infectious cycle is referred to as a passage, and each viral population was carried through ten passages. Following each passage, each viral population was titered, visually examined for plaque morphology (see ‘Materials and Methods’ section for details), and prepared for sequencing. The entire series of 10 passages was performed in triplicate for the Mixed population and once for the Purified population. All replicates were titered and quantified for plaque morphology. Deep sequencing was performed on one lineage for each starting viral population. Plaque images were obtained by plating virus at limiting dilution on monolayers of Vero cells, then fixing and staining with methylene blue at 72 hpi. Tiled images (6 × 6) were exported from Nikon NIS-Elements software, and contrast inverted using Adobe Photoshop to show plaques more clearly. Scale bars indicate 2 mm.