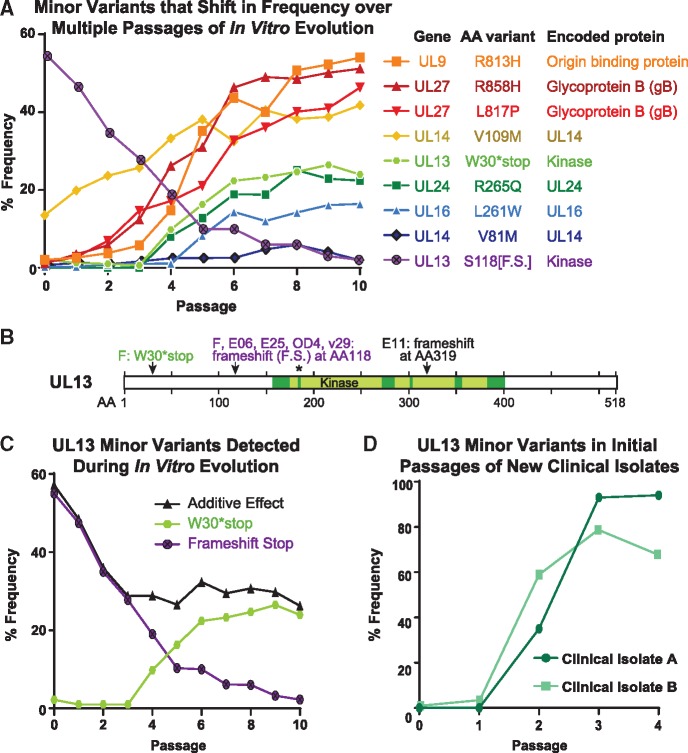
Figure 5.
Minor variant dynamics shift over sequential passages in vitro. (A) A subset of the observed high-confidence minor variants in the Mixed population were plotted by their frequency in the viral population over passage (see Table 1 for full list of minor variants). Each variant would cause a change in the translated protein, whether through premature stop (UL13 S118[F.S.], W30*stop) or a missense variant (all others). Variants and their encoded proteins are listed in the legend according to their frequency at passage 10 (P10). (B) A diagram of the UL13 protein (518 AA in length) depicts the location of the six previously described catalytic domains of this kinase (Smith & Smith 1989), with an asterisk (*) denoting the catalytic lysine where a single point mutation can disrupt kinase activity (Cano-Monreal et al. 2008; Kawaguchi et al. 2003). The minor variants observed in this study, and consensus-level mutations observed in other virus strains in prior studies, are indicated by arrows and labels. GenBank accessions: strain F, GU734771 (Szpara et al. 2010); E25, HM585506; E06, HM585496; E11, HM585500 [all three from (Szpara et al. 2014)]; OD4, JN420342 (Lee et al. 2015). (C) The minor variants observed in the UL13 gene were separated for specific analysis. Each variant is predicted to produce a catalytically-inactive UL13 kinase. Additive Effect refers to the arithmetic addition of the frequency of each of these two individual variants. (D) New clinical isolates of HSV-1 were passaged in Vero cells, genome sequenced, and examined for minor variants in the UL13 gene at each passage. The frequency of genetic variants that would produce an inactive UL13 kinase are plotted over each passage.